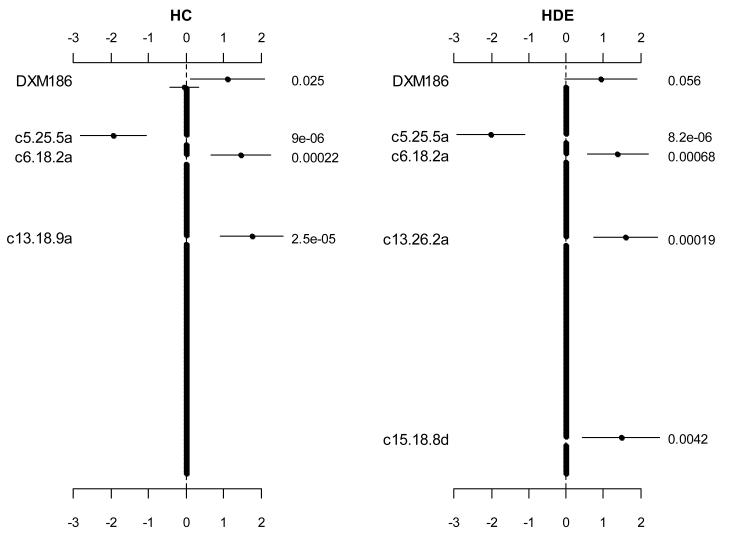
Figure 1.
Jointly fitting two effects of markers (DXM186 and DXM64) on the X chromosome and 262 main effects of 131 markers on 19 autosomes using the hierarchical logistic models with the two prior distributions, hierarchical Cauchy (HC) and hierarchical double-exponential (HDE). The points, short lines and numbers at the right side represent estimates of effects, ± 2 standard errors, and p-values, respectively. Only effects with p-value < 0.06 are labeled. The notation, e.g., c5.25.5a, indicates the additive predictor of the marker located at 25.5 cM on chromosome 5.