Abstract
Purpose of review
The present review discusses the physiological functions of selected caspase recruitment domain (CARD)-containing sensor and adaptor proteins and their role in the pathogenesis of intestinal diseases.
Recent findings
Myeloid and lymphoid cells as well as intestinal epithelial cells express several intracellular CARD-containing proteins. CARD-containing sensors, particularly NOD1 (CARD4), NOD2 (CARD15) and IPAF (CARD12), have an important role in the detection of conserved microbial structures of invading microbial pathogens. Upon ligand recognition and activation, the sensors interact through CARD domains with downstream CARD-containing adaptors including CARD9, RIP2 (CARD3) and ASC (CARD5). Recent data suggest that multiple signaling pathways from Toll-like receptors and non-Toll-receptor pathways converge on these adaptor proteins and that their functions are crucial for the initiation of innate immune responses to invading microbial pathogens.
Summary
CARD-containing adaptors and sensors represent an important family of molecules involved in innate host defense against gastrointestinal pathogens and in the regulation of inflammatory responses, suggesting that further insights into their physiological functions may yield new pharmacological strategies for treating intestinal inflammatory conditions.
Keywords: caspase recruitment domain, innate immunity, intestinal host defense, microbial sensor
Introduction
A single-cell epithelial layer forms a physical barrier between the body and potentially harmful microbes in the intestinal lumen. If the barrier is penetrated, local immune defenses minimize or prevent systemic spread of invading bacteria. Such defenses are commonly divided into adaptive immune responses, with narrow antigen specificity, and innate immune defenses, capable of rapid, broadly specific responses to conserved microbial molecules known as pathogen-associated molecular patterns. Such ‘patterns’ include molecularly diverse components present in bacteria, viruses, parasites and fungi such as lipopolysaccharide, bacterial lipoproteins, flagellin, zymosan, and certain nucleic acids. In contrast to adaptive immunity, the recognition capacity of which is based on millions of different antigen receptors, microbial sensing by the innate immune system is mediated by a much smaller number of receptors, called pattern recognition receptors (PRRs).
Upon recognition of a pathogenic microbial motif by PRRs, multiple downstream signaling cascades are activated through a limited set of adaptor molecules. The activation of mitogen-activated protein kinases (MAPK) and key transcription factors such as nuclear factor-κB and activator protein-1 leads to secretion of cytokines with chemotactic and other proinflammatory functions, and thereby the recruitment of inflammatory cells to the site of infection as a first-line defense against further microbial invasion.
In the past decade many PRRs have been discovered and characterized. The first family of PRRs widely studied was the Toll-like receptor (TLR) family. TLRs are membrane-anchored proteins that sense microbial components with their leucine-rich repeats domain exposed to the extracellular space or the lumen of cellular vacuoles that can take up extracellular materials. Upon activation, TLRs stimulate downstream signaling molecules with their cytoplasmic Toll/IL-1 receptor domain.
A second important group of PRRs, termed nucleotide-binding and oligomerization domain (NOD) proteins or NOD-like receptors (NLRs), is located primarily in the cytosol and also functions in the recognition of conserved microbial products. The NLR family, consisting of more than 20 proteins to date, is characterized by three structural domains: a C-terminal leucine-rich repeats domain involved in ligand recognition, a central NOD domain that plays a role in oligomerization and activation, and an N-terminal effector domain. The latter, which can comprise a pyrin domain, a caspase recruitment domain (CARD), or a baculovirus inhibitor-of-apoptosis repeat domain, enables protein–protein interactions and initiates signal transduction cascades that lead to expression of inflammation-associated genes.
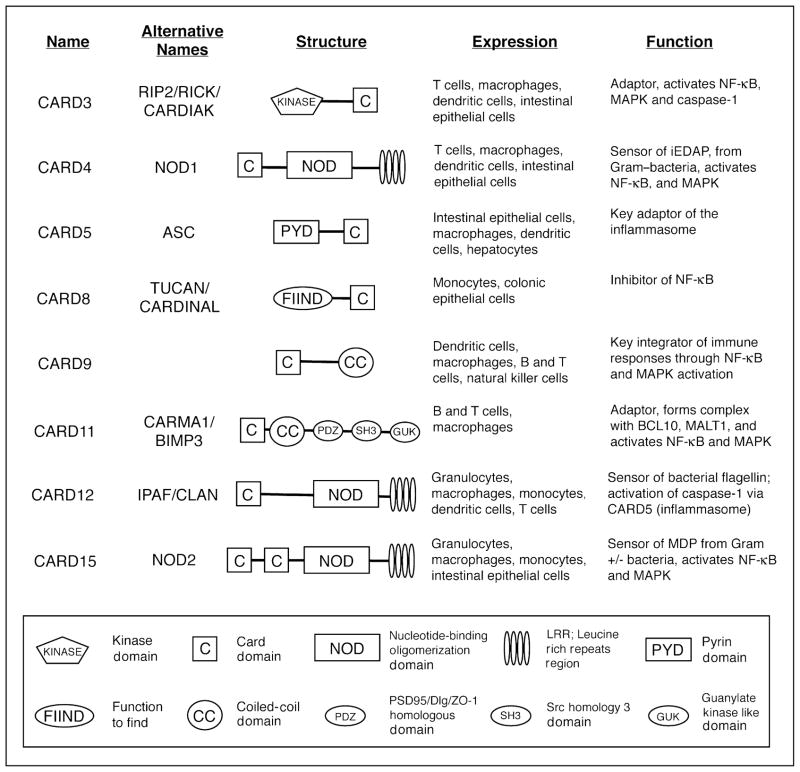
The present review focuses on new developments that demonstrate the importance of selected CARD-containing sensors and adaptor proteins in the pathogenesis of intestinal diseases (Fig. 1). The reader is referred to excellent recent review articles [1,2] that provide comprehensive overviews of the biology of NLRs.
Figure 1. Caspase recruitment domain (CARD)-containing intracellular sensor and adaptor proteins.
The scheme depicts the different CARD-containing adaptors and sensors discussed in the review. CARD6/CARD7/CARD10/CARD14 and other CARD-containing molecules (e.g. caspases, APAF1, CIITA, Arc, RAIDD, ICEBERG) are not included here, as little is known about their intestinal functions. iEDAP, γ-D-glutamyl-meso-diaminopimelic acid; MAPK, mitogen-activated protein kinases; MDP, muramyl dipeptide; NF, nuclear factor.
CARD9: a central adaptor of innate immune signaling pathways
CARD9 is an intracellular adaptor molecule that consists of an N-terminal CARD domain and a C-terminal coiled-coil domain [3] (Fig. 1). It is mainly expressed at the organ level in the bone marrow, thymus, spleen, lung, and liver, and at the cellular level in myeloid dendritic cells and macrophages, but only at low levels in normal T lymphocytes and B lymphocytes [3,4••]. CARD9 expression in the small intestine and colon is low under normal conditions [3], but is probably increased in the course of mucosal inflammation due to an influx of myeloid cells. Originally identified as a binding partner of BCL10, a CARD-containing signaling protein involved in the development of B-cell lymphomas of mucosa-associated lymphoid tissues [3], CARD9 was recently shown to be a key integrator of innate and adaptive immune responses [4••–6••]. Most importantly, mice lacking CARD9 exhibit increased susceptibility to the enteric bacterial pathogen Listeria monocytogenes [4••,5••], as well the fungal pathogen Candida albicans [6••], yet have normal T-lymphocyte and B-lymphocyte development [5••]. The exact mechanisms by which CARD9 mediates effective host defense against different microbial threats are not fully understood, but this adaptor is involved in signaling from a number of innate cell surface receptors, including TLRs, NLRs, and immunoreceptor tyrosine-based activation motifs (ITAM)-associated receptors (Fig. 2).
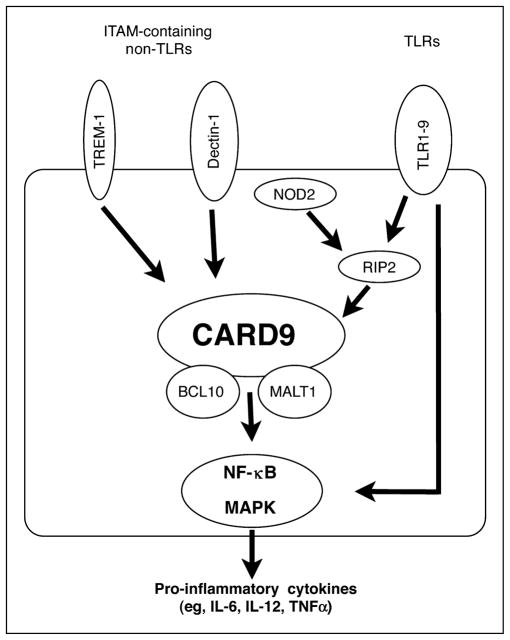
Figure 2. CARD9 functions in myeloid cells.
After activation of membrane-bound microbial sensors such as Toll-like receptors (TLRs) and immunoreceptor tyrosine-based activation motif (ITAM)-containing non-TLRs, and the cytoplasmic sensor NOD2 (CARD15), signaling pathways converge on CARD9. Stimulation of dectin-1 with the fungus-derived ligand zymosan or of TREM-1 (unknown specific ligand) induces phosphorylation of specific tyrosine residues in the ITAMs, leading to downstream formation of a signaling complex of CARD9 with other adaptor proteins, particularly BCL10 and MALT1. TLRs can activate RIP2 (CARD3) and CARD9 in response to a broad range of microbial ligands, while NOD2 interacts with RIP2 and CARD9 via CARD–CARD interactions upon intracellular recognition of muramyl dipeptide. Formation of the signaling complex with CARD9, BCL-10 and MALT1 leads to activation of nuclear factor (NF)-κB and mitogen-activated protein kinases (MAPK), and subsequently secretion of proinflammatory cytokines. TLRs can also activate NF-κB through a CARD9-independent pathway.
CARD9 is required for signaling from most TLRs in dendritic cells, since activation of nuclear factor-κB and/or MAPK, and induction of the inflammatory cytokines TNFα and IL-6, in response to different TLR ligands such as lipopolysaccharide (TLR4), Pam3CSK4 and zymosan (TLR2), flagellin (TLR5), CpG (TLR9), poly(I:C) (TLR3), and loxoribine (TLR7) was defective in CARD9-deficient bone-marrow-derived dendritic cells [4••–6••]. CARD9 may also be required for TLR2/TLR3/TLR7 signaling in macrophages [4••], although another study found only a limited role for CARD9 in TLR7 signaling [5••]. Furthermore, CARD9 mediates signaling by the intracellular microbial sensor NOD2, since CARD9-deficient macrophages failed to increase IL-6 production and/or MAPK activation in response to the NOD2 activator muramyl dipeptide or to infection with the intracellular bacteria L. monocytogenes [4••] (Fig. 2). CARD9 associates with NOD2, as well as the intermediate kinase RIP2 (also termed RICK or CARD3), upon over-expression and/or L. monocytogenes infection [4••], further supporting a role of CARD9 in NOD2 signaling.
Beyond TLRs and NLRs, CARD9 has been shown to be a key adaptor in innate immune signaling by dectin-1 [6••], a sensor of fungal cell wall components and a prototype of innate non-TLRs containing ITAMs [7•,8•]. Other prominent ITAM-containing receptors are FcRγ, which associates with and mediates signaling through several Fc receptors, and DAP12, which associates with the triggering receptor expressed on myeloid cells (TREM) family of receptors and others. Activation of such receptors induces phosphorylation of specific tyrosine residues in the ITAMs, which leads to downstream formation of a signaling complex of CARD9 with other adaptor proteins, particularly BCL10 and MALT1, and activation of nuclear factor-κB and MAPK [5••] (Fig. 2). ITAM-containing receptors are found on lymphocytes, natural killer cells, and myeloid cells, including macrophages, neutrophils, mast cells, and dendritic cells. The importance of CARD9 in signaling through ITAM-containing receptors is demonstrated by the findings that CARD9-deficient macrophages and/or dendritic cells have impaired cytokine production in response to zymosan stimulation of dectin-1, or after crosslinking of FcRγ or of several DAP12-associated receptors [5••]. Consequently, host defense against C. albicans, which may partly depend on dectin-1 [7•], is compromised in CARD9-deficient mice [6••]. The underlying mechanism may involve a role of CARD9 in the development of IL-17 producing CD4 T-cell effectors (Th17 cells), which are induced in a CARD9-dependent manner upon C. albicans infection [9•].
Among the TREM family members, whose signaling depends on CARD9, TREM-1 has garnered recent interest in intestinal immunology as a regulator of mucosal inflammation. TREM-1 is expressed constitutively in neutrophils and monocytes/macrophages, and is further induced in these cells by stimulation with microbial products (e.g. lipopolysaccharide and lipoteichoic acid) and exposure to extracellular bacteria such as Pseudomonas aeruginosa and Staphylococcus aureus [10,11]. Activation of TREM-1 by crosslinking with monoclonal antibodies leads to secretion of proinflammatory chemokines, especially when TLR or NLR ligands are used as a costimulus, indicating that TREM-1 can amplify proinflammatory responses induced by TLRs or NLRs [12,13••]. It is presently unclear whether TREM-1 recognizes a specific microbial ligand. TREM-1-expressing macrophages are significantly increased in the inflamed mucosa of patients with inflammatory bowel disease and in mouse models of colitis [13••], whereas little TREM-1 expression is found on resident macrophages of the normal human small intestine or colon [11]. Furthermore soluble TREM-1 levels were elevated in the serum of inflammatory bowel disease patients [14]. Importantly, specific blocking of TREM-1 with an antagonistic peptide attenuated inflammation in mouse colitis models [13••]. These data indicate that this ITAM-containing receptor contributes to mucosal inflammation, and suggests more broadly that CARD9-dependent signaling processes promise to be valuable targets in the treatment of inflammatory bowel disease.
CARD4 (NOD1) and CARD15 (NOD2): cytoplasmic sensors of bacterial peptidoglycans
The NLR proteins NOD1 (encoded by CARD4) and NOD2 (encoded by CARD15) have a tripartite structure with one (NOD1) or two (NOD2) N-terminal CARD domains, a centrally located NOD domain, and a C-terminal set of leucine-rich repeats involved in ligand recognition (Fig. 1). NOD1 is expressed in many cell types, while NOD2 is constitutively expressed predominantly in myeloid cells (neutrophils, macrophages, dendritic cells) and in Paneth cells of the small intestine [15,16]. Both proteins are cytosolic sensors of peptidoglycans, components of bacterial cell walls. NOD1 senses the peptidoglycan-derived peptide γ-D-glutamyl-meso-diaminopimelic acid, which is present mainly in Gram-negative bacteria [17,18]. NOD2 detects muramyl dipeptide, which can be found in a wide range of both Gram-positive and Gram-negative bacteria [19], indicating that its recognition specificity is markedly broader than that of NOD1.
NOD1 contributes to recognition of several enteropathogenic bacteria including Shigella flexneri [20], enteroinvasive Escherichia coli [21], Helicobacter pylori [22], and Campylobacter jejuni [23], while NOD2 was shown to be involved in innate detection of L. monocytogenes and Salmonella typhimurium [24–26]. Several of these pathogens (e.g. S. flexneri, L. monocytogenes) invade host cells and reside in the cytoplasm, where they may release cell wall components that can make contact with cytoplasmic NOD1 or NOD2. Others (C. jejuni, S. typhimurium) are invasive but are not known to enter the cytoplasm of host cells, or are only minimally invasive (H. pylori), which raises questions about the exact mechanisms by which microbe-derived peptidoglycans are delivered into the cytosol and are sensed by NOD1 or NOD2. Intracellular peptidoglycan delivery can occur via phagolysosomes or via specialized bacterial secretion systems [22,27], or by subversion of peptide transport pathways such as the intestinal peptide transporter hPEPT [28]. Upon activation, NOD1 and NOD2 associate through CARD–CARD interactions with the CARD-containing serine/threonine kinase RIP2 (CARD3), which leads to activation of nuclear factor-κB and MAPK, and consequently to secretion of proinflammatory cytokines [24,25].
Mice lacking NOD1 or NOD2 are generally healthy and fertile [17,29,30•], indicating that these sensors have no critical developmental functions. A recent report showed that the Peyer’s patches of conventionally reared NOD2-deficient mice had elevated numbers of CD4+ T cells and M cells, as well as increased expression of proinflammatory cytokines [30•]. Oral administration of Saccharomyces cerevisiae or killed E. coli or S. aureus led to increased bacterial translocation across Peyer’s patches, suggesting that NOD2 deficiency causes an epithelial barrier defect. Furthermore, rectal administration of the colitis-inducing agent trinitrobenzene sulphonic acid caused elevated proinflammatory cytokine production in the colon of NOD2-deficient mice [30•], which suggests that loss of normal NOD2 functions can exacerbate colitis induced by inflammatory agents. This notion is further supported by recent work in NOD2 transgenic mice engineered to overexpress normal murine NOD2 in antigen-presenting and other cells under the control of a major histocompatibility complex class II promoter [31••]. These mice exhibited decreased mucosal inflammation and proinflammatory cytokine production in two different colitis models, consistent with the idea that normal NOD2 suppresses mucosal inflammatory responses. A similar attenuation of colitis was observed when wild-type mice were injected with an encapsulated NOD2 expression plasmid [31••]. Interestingly, injection with a plasmid encoding a mutant form of mouse NOD2 equivalent to NOD21007fs associated with Crohn’s disease caused less attenuation of colitis, suggesting that the mutation interfered with a normal suppressive function of NOD2 [31••]. In this context, NOD2 has been shown to inhibit signaling through the TLR2 pathway [32•], although TLR2 can also protect against colitis under certain conditions [33,34]. The relative roles of TLR2 and NOD2 in regulating mucosal inflammation of different etiologies, including Crohn’s disease, remain to be fully understood [35].
Beyond studies on the role of NOD1/NOD2 in regulating inflammatory processes, several reports have addressed NOD1/NOD2-dependent host defense against specific microbial threats. For example, NOD2-deficient mice infected with invasive L. monocytogenes showed increased bacterial numbers in the spleen when challenged orally but not after intraperitoneal injection, indicating that NOD2 had a physiological function in local intestinal defense against the bacteria [24]. Expression of several genes encoding for cryptdins (peptides with antimicrobial activity produced by intestinal Paneth cells) was decreased upon infection of NOD2-deficient mice with Listeria, suggesting that NOD2 can regulate innate antibacterial defenses in the intestinal tract. Consistent with this concept, if not the actual mechanisms, intestinal epithelial cells engineered to overexpress NOD2 had reduced numbers of intracellular bacteria upon infection with S. typhimurium [26]. In regard to NOD1, mice lacking this sensor were shown to be significantly more susceptible to oral infection with H. pylori [22]. A possible underlying mechanism is suggested by the finding that primary gastric epithelial cells of NOD1-deficient mice produced lower levels of the neutrophil chemoattractant MIP-2 (CXCL2) upon stimulation with H. pylori [22]. Diminished epithelial chemokine production may compromise effector cell recruitment and thereby host defense against the bacteria.
CARD12 (IPAF/CLAN) and CARD5 (ASC): components of the inflammasome
IPAF is a CARD-containing NLR with a tripartite structure similar to NOD1 (Fig. 1) and serves likewise as a cytoplasmic sensor of microbial components. IPAF appears to be a cytosolic counterpart of TLR5, as it detects intracellular bacterial flagellin, a protein required for bacterial motility, from the enteropathogen S. typhimurium and from Legionella pneumophila, causative agent of Legionnaire’s disease in the lungs [36•]. The dysentery-causing enteropathogen S. flexneri is also detected by IPAF, but, unlike Salmonella and Legionella, flagellin is dispensable in Shigella recognition [37]. Upon detection of these invasive pathogens, IPAF forms a complex with the key inflammasome adaptor apoptosis-associated speck-like protein (ASC/CARD5), which leads to caspase-1 activation, and to processing and subsequent release of mature IL-1β and IL-18. IPAF-deficient macrophages did not process or release IL-1β upon stimulation with S. typhimurium, while they were not defective in IL-1β processing and release in response to TLR agonists plus ATP [38,39]. Similarly, activation of caspase-1 in response to S. typhimurium was abrogated in ASC-deficient macrophages [38]. These data indicate that IPAF (via ASC) mediates the recognition of a narrow spectrum of microbial triggers. Besides its specific role in the activation of caspase-1 and IL-1β release, IPAF was also reported to inhibit nuclear factor-κB signaling, since its overexpression in HEK 293 cells suppressed NOD1-dependent and NOD2-dependent nuclear factor-κB activation [40]. Inhibition was accompanied by association of IPAF with NOD1 and NOD2 upon peptidoglycan stimulation in the human monocytic cell line THP1 [40], suggesting that hetero-oligomerization between different NOD domain-containing signal adaptors modulates innate immune response to bacteria and their products.
CARD8 (TUCAN/CARDINAL): negative regulator of nuclear factor-κB signaling
At least two CARD-containing adaptors, CARD8 and CARD6, have been identified with regulatory and inhibitory functions in nuclear factor-κB signaling and/or inhibition of IL-1β processing. One of the adaptors, CARD8 (TUCAN/CARDINAL), mainly expressed in monocytes/macrophages, is an adaptor protein that inhibits TNFα and RIP2-induced nuclear factor-κB activation [41,42]. The adaptor may bind directly to the regulatory subunit of the IκB kinase complex, IκB kinase γ (NEMO), and inhibit IκB kinase activation, although another study did not observe such an association [42]. In addition, CARD8 also interacts with and negatively regulates caspase-1, thereby suppressing release of mature IL-1β, and inhibits caspase-9 activation and apoptosis [42]. Interestingly, an association between a genetic CARD8 variant (nonsense mutation rs2043211; stop codon at Cys10) and Crohn’s disease has been suggested in one recent report [43], although several other studies in British, German and Norwegian populations [44,45] did not find a significant association between genetic CARD8 polymorphisms and Crohn’s disease, and thus concluded that this gene is not a susceptibility locus for this condition. Beyond its role in inflammation, CARD8 may contribute to colon cancer pathogenesis. Increased CARD8 expression was found by immunostaining in colon cancers compared with adjacent unaffected colonic tissue [46]. Overexpression of CARD8 in epithelial cell lines protected the cells against inducible apoptosis [46], suggesting that this adaptor can contribute to enhanced survival of colon cancer cells by inhibiting normal epithelial cell death.
Conclusion
The mucosal immune system is confronted with an ongoing microbial challenge due to the presence of an abundant commensal microbiota and the intermittent exposure to microbial pathogens in food and water. Intra-cellular CARD-containing sensor and adaptor proteins are able to detect conserved structures present in gastrointestinal and other microbes and to initiate inflammatory responses, making them important players in innate defense against a wide range of invading microbial pathogens. The central immunologic roles of these sensors and adaptors suggest that they will be prime targets in the rational design of novel vaccine-based prevention strategies against gastrointestinal infections, and in the treatment of chronic inflammatory diseases in the intestine. Full exploitation of the CARD-carrying sensors and adaptors as targets of immunomodulatory intervention strategies, however, will require further characterization of their full physiologic functions under normal and disease conditions.
Acknowledgments
The work was supported by a research fellowship from the Swiss National Foundation to P.H. (SSMBS; PASMA 114623), and NIH grants AI56075, DK70867, DK80506, RR17030, and DK35108. The authors thank Gregory Botwin for help with the manuscript preparation.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest Additional references related to this topic can also be found in the Current World Literature section in this issue (pp. 237–238).
- 1.Fritz JH, Ferrero RL, Philpott DJ, et al. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- 2.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 3.Bertin J, Guo Y, Wang L, et al. CARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-kappa B. J Biol Chem. 2000;275:41082–41086. doi: 10.1074/jbc.C000726200. [DOI] [PubMed] [Google Scholar]
- 4••.Hsu YM, Zhang Y, You Y, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8:198–205. doi: 10.1038/ni1426. This important paper, along with references [5••,6••], demonstrates for the first time the central relevance of the adaptor CARD9 in integrating signals from TLRs and NOD microbial sensors for the induction of inflammatory mediators. It further shows the physiological importance of CARD9 in host defense against the model bacterial pathogen Listeria. [DOI] [PubMed] [Google Scholar]
- 5••.Hara H, Ishihara C, Takeuchi A, et al. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat Immunol. 2007;8:619–629. doi: 10.1038/ni1466. This work, in parallel with references [4••,6••], shows that CARD9 integrates signals from TLR and non-TLR immunoreceptors, and thereby forms a key conduit for the integration of innate immune recognition systems. [DOI] [PubMed] [Google Scholar]
- 6••.Gross O, Gewies A, Finger K, et al. Card9 controls a non-TLR signalling pathway for innate antifungal immunity. Nature. 2006;442:651–656. doi: 10.1038/nature04926. This paper, along with references [4••,5••], provides important new insights into the role of the central adaptor CARD9 in the function of innate immune recognition of fungi. Further experiments demonstrate convincingly that CARD9 is required for effective host defense against fungal pathogens. [DOI] [PubMed] [Google Scholar]
- 7•.Taylor PR, Tsoni SV, Willment JA, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol. 2007;8:31–38. doi: 10.1038/ni1408. The study defines the physiological importance of dectin-1 as an innate receptor for recognition of the fungal cell wall component β-glucan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Saijo S, Fujikado N, Furuta T, et al. Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans. Nat Immunol. 2007;8:39–46. doi: 10.1038/ni1425. This work, along with reference [7•], underlines the importance of dectin-1 for innate defense against fungal pathogens. [DOI] [PubMed] [Google Scholar]
- 9•.LeibundGut-Landmann S, Gross O, Robinson MJ, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–638. doi: 10.1038/ni1460. The data in this paper prove that CARD9 is involved in coupling innate and adaptive immunity as shown by CARD9-dependent induction of T helper 17 cells upon infection with C. albicans. [DOI] [PubMed] [Google Scholar]
- 10.Bouchon A, Facchetti F, Weigand MA, et al. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410:1103–1107. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- 11.Schenk M, Bouchon A, Birrer S, et al. Macrophages expressing triggering receptor expressed on myeloid cells-1 are underrepresented in the human intestine. J Immunol. 2005;174:517–524. doi: 10.4049/jimmunol.174.1.517. [DOI] [PubMed] [Google Scholar]
- 12.Netea MG, Azam T, Ferwerda G, et al. Triggering receptor expressed on myeloid cells-1 (TREM-1) amplifies the signals induced by the NACHT-LRR (NLR) pattern recognition receptors. J Leukoc Biol. 2006;80:1454–1461. doi: 10.1189/jlb.1205758. [DOI] [PubMed] [Google Scholar]
- 13••.Schenk M, Bouchon A, Seibold F, et al. TREM-1-expressing intestinal macrophages crucially amplify chronic inflammation in experimental colitis and inflammatory bowel diseases. J Clin Invest. 2007;117:3097–3106. doi: 10.1172/JCI30602. In this study, the importance of TREM-1 for inflammatory responses in the intestine is highlighted. TREM-1 expression was upregulated in patients with IBD and in experimental mouse models of colitis. Specific blocking of TREM-1 with an antagonistic peptide attenuated inflammation in murine colitis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tzivras M, Koussoulas V, Giamarellos-Bourboulis EJ, et al. Role of soluble triggering receptor expressed on myeloid cells in inflammatory bowel disease. World J Gastroenterol. 2006;12:3416–3419. doi: 10.3748/wjg.v12.i21.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogura Y, Lala S, Xin W, et al. Expression of NOD2 in Paneth cells: a possible link to Crohn’s ileitis. Gut. 2003;52:1591–1597. doi: 10.1136/gut.52.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lala S, Ogura Y, Osborne C, et al. Crohn’s disease and the NOD2 gene: a role for Paneth cells. Gastroenterology. 2003;125:47–57. doi: 10.1016/s0016-5085(03)00661-9. [DOI] [PubMed] [Google Scholar]
- 17.Chamaillard M, Hashimoto M, Horie Y, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 18.Girardin SE, Boneca IG, Carneiro LA, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 19.Inohara N, Ogura Y, Fontalba A, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 20.Girardin SE, Tournebize R, Mavris M, et al. CARD4/Nod1 mediates NF-κB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2001;2:736–742. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim JG, Lee SJ, Kagnoff MF. Nod1 is an essential signal transducer in intestinal epithelial cells infected with bacteria that avoid recognition by toll-like receptors. Infect Immun. 2004;72:1487–1495. doi: 10.1128/IAI.72.3.1487-1495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viala J, Chaput C, Boneca IG, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 23.Zilbauer M, Dorrell N, Elmi A, et al. A major role for intestinal epithelial nucleotide oligomerization domain 1 (NOD1) in eliciting host bactericidal immune responses to Campylobacter jejuni. Cell Microbiol. 2007;9:2404–2416. doi: 10.1111/j.1462-5822.2007.00969.x. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 25.Park JH, Kim YG, McDonald C, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol. 2007;178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- 26.Hisamatsu T, Suzuki M, Reinecker HC, et al. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology. 2003;124:993–1000. doi: 10.1053/gast.2003.50153. [DOI] [PubMed] [Google Scholar]
- 27.Herskovits AA, Auerbuch V, Portnoy DA. Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS Pathog. 2007;3:e51. doi: 10.1371/journal.ppat.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vavricka SR, Musch MW, Chang JE, et al. hPepT1 transports muramyl dipeptide, activating NF-κB and stimulating IL-8 secretion in human colonic Caco2/bbe cells. Gastroenterology. 2004;127:1401–1409. doi: 10.1053/j.gastro.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 29.Pauleau AL, Murray PJ. Role of nod2 in the response of macrophages to toll-like receptor agonists. Mol Cell Biol. 2003;23:7531–7539. doi: 10.1128/MCB.23.21.7531-7539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Barreau F, Meinzer U, Chareyre F, et al. CARD15/NOD2 is required for Peyer’s patches homeostasis in mice. PLoS ONE. 2007;2:e523. doi: 10.1371/journal.pone.0000523. This study describes abnormal functions of Peyer’s patches in NOD2-deficient mice, which exhibited exaggerated immune responses and increased bacterial translocation across Peyer’s patches. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Yang Z, Fuss IJ, Watanabe T, et al. NOD2 transgenic mice exhibit enhanced MDP-mediated down-regulation of TLR2 responses and resistance to colitis induction. Gastroenterology. 2007;133:1510–1521. doi: 10.1053/j.gastro.2007.07.025. In this provocative and important study, novel transgenic mice were generated that overexpress murine NOD2 to demonstrate that NOD2 inhibits signaling through TLR2. Furthermore, transgenic mice had attenuated mucosal inflammation in two different colitis models, underlining the potential physiological relevance of the signaling observations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Watanabe T, Kitani A, Murray PJ, et al. Nucleotide binding oligomerization domain 2 deficiency leads to dysregulated TLR2 signaling and induction of antigen-specific colitis. Immunity. 2006;25:473–485. doi: 10.1016/j.immuni.2006.06.018. This paper, along with reference [31••], demonstrates for the first time that normal NOD2 can inhibit signaling through TLR2, while mutant NOD2 failed to do so, suggesting a mechanism by which Crohn’s disease-associated NOD2 proteins might exacerbate mucosal inflammation by failing to downregulate proinflammatory TLR2 signaling. [DOI] [PubMed] [Google Scholar]
- 33.Gibson DL, Ma C, Rosenberger CM, et al. Toll-like receptor 2 plays a critical role in maintaining mucosal integrity during Citrobacter rodentium-induced colitis. Cell Microbiol. 2008;10:388–403. doi: 10.1111/j.1462-5822.2007.01052.x. [DOI] [PubMed] [Google Scholar]
- 34.Cario E, Gerken G, Podolsky DK. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology. 2007;132:1359–1374. doi: 10.1053/j.gastro.2007.02.056. [DOI] [PubMed] [Google Scholar]
- 35.Eckmann L, Karin M. NOD2 and Crohn’s disease: loss or gain of function? Immunity. 2005;22:661–667. doi: 10.1016/j.immuni.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 36•.Franchi L, Amer A, Body-Malapel M, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in Salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. This work defines IPAF as an intracellular receptor for bacterial flagellin, making it only the second innate sensor (besides TLR5) that can recognize this important bacterial protein. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki T, Franchi L, Toma C, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mariathasan S, Newton K, Monack DM, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 39.Mariathasan S, Monack DM. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat Rev Immunol. 2007;7:31–40. doi: 10.1038/nri1997. [DOI] [PubMed] [Google Scholar]
- 40.Damiano JS, Oliveira V, Welsh K, et al. Heterotypic interactions among NACHT domains: implications for regulation of innate immune responses. Biochem J. 2004;381:213–219. doi: 10.1042/BJ20031506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouchier-Hayes L, Conroy H, Egan H, et al. CARDINAL, a novel caspase recruitment domain protein, is an inhibitor of multiple NF-κB activation pathways. J Biol Chem. 2001;276:44069–44077. doi: 10.1074/jbc.M107373200. [DOI] [PubMed] [Google Scholar]
- 42.Razmara M, Srinivasula SM, Wang L, et al. CARD-8 protein, a new CARD family member that regulates caspase-1 activation and apoptosis. J Biol Chem. 2002;277:13952–13958. doi: 10.1074/jbc.M107811200. [DOI] [PubMed] [Google Scholar]
- 43.McGovern DP, Butler H, Ahmad T, et al. TUCAN (CARD8) genetic variants and inflammatory bowel disease. Gastroenterology. 2006;131:1190–1196. doi: 10.1053/j.gastro.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 44.Franke A, Rosenstiel P, Balschun T, et al. No association between the TUCAN (CARD8) Cys10Stop mutation and inflammatory bowel disease in a large retrospective German and a clinically well characterized Norwegian sample. Gastroenterology. 2007;132:2080–2081. doi: 10.1053/j.gastro.2007.03.087. [DOI] [PubMed] [Google Scholar]
- 45.Fisher SA, Mirza MM, Onnie CM, et al. Combined evidence from three large British Association studies rejects TUCAN/CARD8 as an IBD susceptibility gene. Gastroenterology. 2007;132:2078–2080. doi: 10.1053/j.gastro.2007.03.086. [DOI] [PubMed] [Google Scholar]
- 46.Pathan N, Marusawa H, Krajewska M, et al. TUCAN, an antiapoptotic caspase-associated recruitment domain family protein overexpressed in cancer. J Biol Chem. 2001;276:32220–32229. doi: 10.1074/jbc.M100433200. [DOI] [PubMed] [Google Scholar]