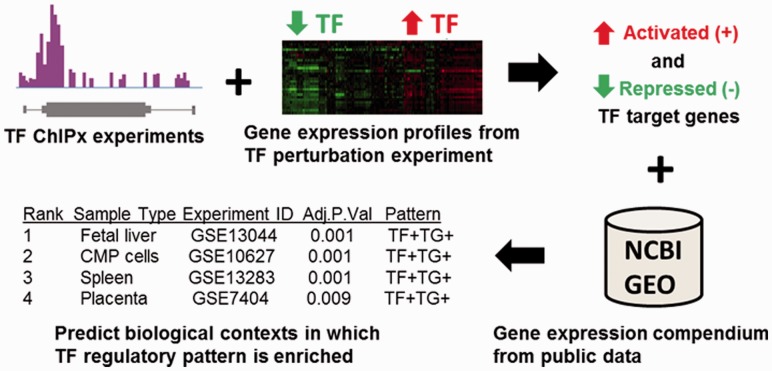
Fig. 1.
ChIP-PED overview. Gene expression profiles from TF perturbation experiments are intersected with ChIPx experiments to obtain a set of activated and repressed target genes. ChIP-PED then takes as input the TF and target genes and scans through a compendium of publicly available gene expression profiles to search for biological contexts in which the TF and target genes are enriched in activity. The final output is a ranked table of biological contexts enriched with a regulatory pattern of interest