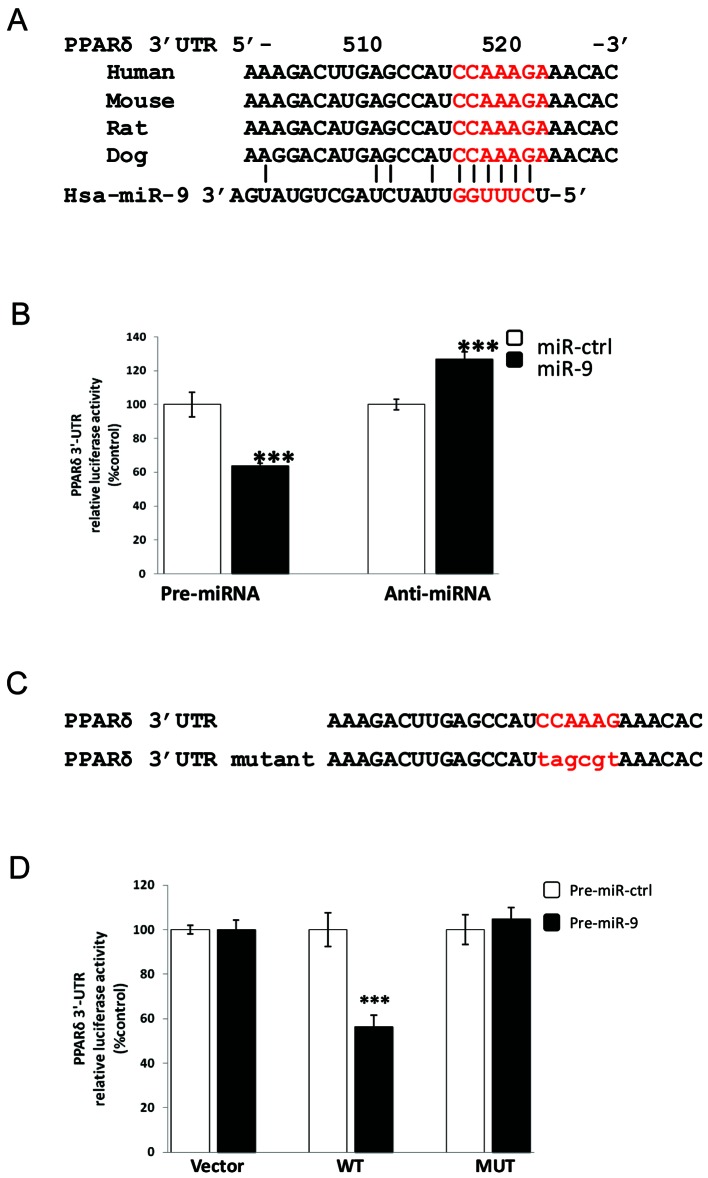
Figure 1.
PPARδ is the direct target of miR-9. (A) DNA sequence alignment of the evolutionary conserved miR-9 binding site in the PPARδ 3′-UTR of mammals. The miR-9 seed match region is highlighted in red capital letters. (B) The relative expression obtained from reporter constructs containing the wild-type (WT) PPARδ 3′-UTR that were transfected into HEK293 cells together with pre-miR-9 or miR-9 inhibitor oligonucleotide (anti-miR-9). The luciferase activity was measured 24 h after transfection. (C) The nucleotides changed in the PPARδ 3′-UTR mutant construct are indicated in red lower case. (D) The relative expression obtained when HEK293 cells were transfected with reporter constructs containing wild-type (WT) or mutant (MUT) PPARδ 3′-UTR or empty renilla expression vector (vector) together with miR-9 precursor RNA (pre-miR-9) or miRNA precursor control (pre-miR-ctrl). The luciferase activity was measured 24 h after transfection. The experiments were repeated 3 times. A representative result is shown. Each bar represents the mean ± SD for 3 wells. ***P<0.001.