Abstract
Using a collection of dye-labeled cytochrome c (cyt c) variants, we identify transformations of heterogeneous cardiolipin (CL)-bound cyt c ensemble with added ATP. Distributions of dye-to-heme distances P(r) from time-resolved FRET show that ATP decreases the population of largely unfolded cyt c conformers, but its effects are distinct from those of a simple salt. High peroxidase activity of CL-bound cyt c with added ATP suggests binding interactions that favor protein structures with the open heme pocket. Although ATP weakens cyt c – CL binding interactions, it also boosts the apoptosis-relevant peroxidase activity of CL-bound cyt c.
Adenosine 5′-triphosphate (ATP) and cytochrome c (cyt c) are among the major components of mitochondria1 and their interactions have been subjects of great interest.2,3 Similar to other ionic compounds,4 ATP promotes refolding of several denatured forms of cyt c to globular structures.5 Recent studies suggest an intriguing role for ATP in regulating cyt c-cardiolipin (CL) interactions. A critical interaction in apoptosis, cyt c binding to CL membranes unfolds the protein, allowing reactive oxygen species to access the heme and resulting in CL oxidation, that, in turn, facilitates cyt c release into the cytosol.6 High millimolar concentrations of ATP have been shown to dissociate the cyt c-CL complex.7 Furthermore, ATP dramatically reduces cyt c-induced formation of pores in CL membranes.8 However, the structural features of CL-bound cyt c that underlie the mechanisms of these ATP effects are currently unclear.
Cyt c-CL binding is strongly driven by ionic interactions between the positively-charged protein and the negatively-charged lipid.9 Additional ions in solution mask these interactions. Due to the charged nature of ATP, we attempted to separate specific effects of the ATP moiety from any nonspecific ionic effects by comparing cyt c-CL interactions in the presence of added ATP (10 mM) or NaCl (86 mM, salt) at identical ionic strengths. Ultracentrifugation pelleting experiments and biolayer interferometry measurements revealed similar binding behavior under these two experimental conditions (Figure S1), demonstrating that the protein-lipid binding is largely controlled by ionic strength. With similar percentages of the bound protein, these conditions enabled direct comparison of the ATP- and salt-induced conformational ensembles.
CL-bound cyt c is a heterogeneous ensemble of non-native protein species, whose properties are difficult to characterize with traditional structural methods.10 We employed fluorescence and peroxidase activity measurements to shed light on the structural and functional effects of ATP on CL-bound cyt c. Five dansyl (Dns)-labeled variants (Figures 1 and S2) probed multiple sites near the protein C-terminus, the region that is particularly sensitive to protein unfolding.10 These sites are located close to the residues thought to be involved in binding CL (Asn52, Lys72, -73, -86, and 87),11 as well as ATP (Glu62, Lys88, and Arg91)2. All five variants have structures similar to those of the wild-type protein and their stabilities are minimally affected by the modification (Table S1). The heme acts as a quencher of Dns fluorescence, with a critical length R0 of the donor (D)-acceptor (A) pair of 39 Å.
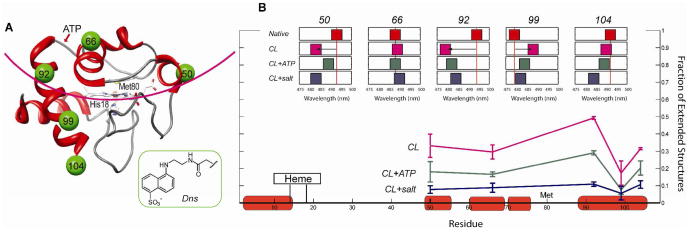
Figure 1.
(A) Structure of the native horse heart cyt c showing the labeled positions and the major ATP binding site (S1); inset –the Dns label. The arc line shows liposome binding surface suggested by results of this study. (B) Dns emission maxima (top) and fractions of E (r≥30 Å) species from TR-FRET (bottom) of five variants (3 μM) in a 25 mM HEPES buffer at pH 7.4; with CL liposomes (50 mol% CL, 750 μM total lipid); with CL liposomes and 10 mM ATP; with CL liposomes and 86 mM salt.
The new set of Dns variants revealed a clear picture of the cyt c binding surface. The large blue shifts of Dns emission maxima (Figure 1B) for Dns50 and Dns92 suggest the proximity of these sites to the lipid, while the red shift for Dns99 places this residue away from the liposome surface and is consistent with the protein unfolding. Time-resolved FRET (TR-FRET) in all variants revealed the coexistence of compact (C) and extended (E) protein conformers. The partitioning between C and E species reflects the rate of the conformational exchange, which depends on the degree of protein unfolding and cyt c-CL interactions.12 The low population of E conformers for the Dns99 variant is consistent with positioning of residue 99 away from the liposome surface and its location close to the critical stabilizing contacts between the N- and C- terminal helices in native cyt c.13 Residue 104, on the other hand, appears to move to a more nonpolar environment upon cyt c binding to CL liposomes; the higher fraction of E species in Dns104, compared to Dns99, supports a suggested fraying of the protein C-helix. Sensitive to unfolding of the Met80-containing loop, the Dns92 variant shows the largest fraction of E species.10
Consistent with the decrease in cyt c-CL affinity, addition of salt or ATP reduces the population of E structures for all five variants. However, a greater fraction of E structures with ATP suggests additional effects of this compound beyond simple charge screening. The labeling site 92 is a particularly strong reporter of distinct effects of ATP and salt on the protein conformational ensemble (Figure 1B).
ATP and GTP similarly affect Dns-heme distance distributions P(r) (Figure S3), suggesting that maintenance of the E structures is not nucleoside specific. The presence of the nucleoside moiety, however, is important since lower populations of E species are observed with just charged polyphosphates. No changes in Dns fluorescence decays are detected upon addition of ATP to the native cyt c variants, in accord with only minor structural perturbations in the ATP adduct of the protein.2 Instead, ATP appears to more dramatically affect conformations of CL-bound cyt c.
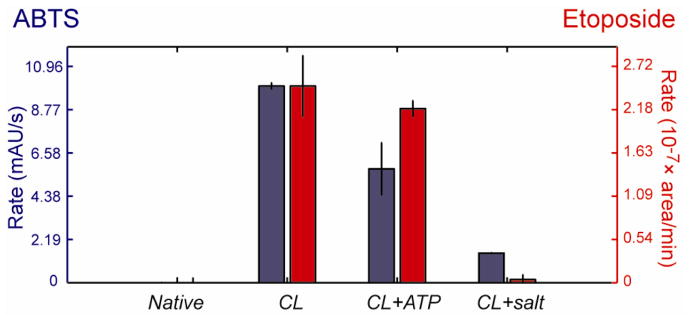
We performed two different peroxidase activity assays to evaluate the functional effects of ATP on the CL-bound cyt c. The peroxidase activity of cyt c is minimal in the native state but increases greatly upon protein binding to CL liposomes (Figures 2 and S4). Addition of salt decreases the peroxidase activity of CL-bound cyt c almost to that of the native protein. Addition of ATP, however, does not change the peroxidase activity much from the levels in CL-bound protein implying retention of the open heme pocket. These results, from two different activity assays, highlight the differences between the two conformational ensembles. Although both salt and ATP inhibit cyt c - liposome interactions, the latter favors highly-reactive protein species.
Figure 2.
Initial rates of 2,2′azinobis-(2-ethylbenzthiazoline-6-sulfonate (ABTS) oxidation ([ABTS]=50 μM and [H2O2]=1 mM) and etoposide phenoxyl radical formation ([etoposide]=100 μM and [H2O2]=100 μM) catalyzed by cyt c (3 μM) in a 25 mM HEPES buffer at pH 7.4; with CL liposomes (50 mol% CL, 750 μM total lipid); with CL liposomes and 10 mM ATP; with CL liposomes and 86 mM salt.
To rationalize these findings, conformational dynamics of CL-bound cyt c in the presence of added ATP or salt were probed by fluorescence correlation spectroscopy (FCS).12,14 Cyt c was labeled with the bright fluorophores nitrobenzoxadiazole (NBD, hydrophobic) and Alexa488 (Alx, hydrophilic) at the identified lipid insertion site at residue 92 and solvent-accessible site at residue 99, respectively. Similar to Dns variants (Figure 1), the fluorescence signals of the dyes are quenched in the native labeled proteins and dramatically increase upon cyt c binding to CL (Figure S5). The blue shift of the NBD emission maxima in CL-bound NBD92 variant confirmed the position of this small dye in the nonpolar environment, with and without added ATP or salt.
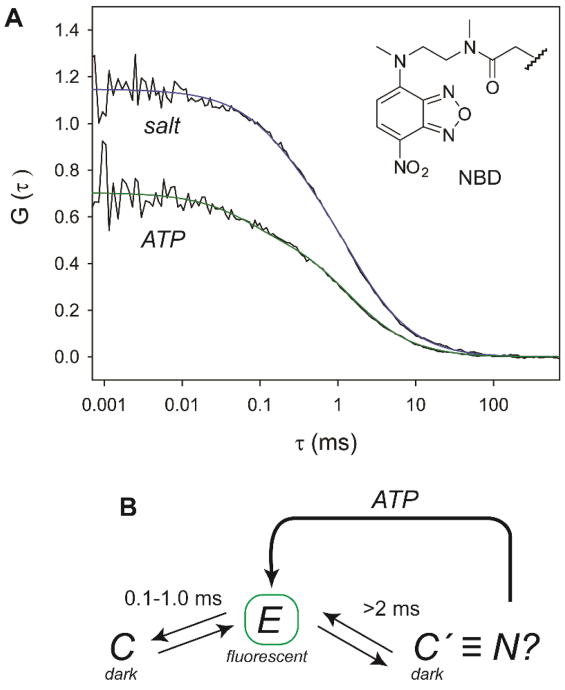
FCS curves (Figure 3), collected at very high lipid-to-protein ratios, provided information on the liposome diffusion times (τd=2 ms) and fast (τex <τd) dynamics associated with exchange between dark C and fluorescent E states in CL-bound cyt c. With added ATP or salt, the exponential relaxation component (τex of 0.8-1.0 ms and ≈100 μs for NBD92 and Alx99, respectively) is clearly apparent in the FCS data (Figures 3 and S6); this conformational exchange is thought to arise from the protein folding into a compact state and depends on the protein interactions with the liposome surface.12 For each of the labeled variants, the FCS curves with added ATP and salt are remarkably alike (Figures 3A and S6), further illustrating similarities of the general ionic effects of the two additives.
Figure 3.
(A) FCS curves for NBD92 cyt c (50 nM) in a 25 mM HEPES buffer at pH 74 with CL liposomes (50 mol% CL, 750 μM total lipid) and 10 mM ATP or 86 mM salt. (B) Scheme showing the species in the conformational ensemble of CL-bound cyt c with added ATP or salt. The fast (0.1-1.0 ms) exchange between compact (C) and extended (E) species is observed directly, while the native-like species C′ is inferred from the analysis of the G(0) values. ATP destabilizes C′ increasing the population of E.
The most interesting result is an increase in the G(0) values for the samples with added salt compared to that with ATP, suggesting a greater population of cyt c molecules in the dark state under these conditions (Figure 3). With very similar FCS curves for the two additives, these differences cannot be explained solely by the species undergoing submillisecond C ⇄ E exchange and instead suggest the presence of another compact species, C′, that likely exchanges slowly (>2 ms) with the highly fluorescent E species. The low peroxidase activity of CL-bound cyt c with added salt hints to a well-protected, possibly native, heme environment in C′. The slow (seconds)15 rate of CL-induced cyt c unfolding (N ⇄ E) is consistent with the timescale limit (>2 ms) for the C′ ⇄ E exchange set by FCS. While both ATP and salt weaken ionic interactions and promote collapse of unfolded cyt c, the two additives affect differently the population of C′.
A simple proposal can explain the effects of ATP on CL-bound cyt c. The major site (S1)2 for ATP binding next to Arg91 in the native cyt c (Figure 1A) includes some of the positively charged residues implicated in CL binding. The decrease in CL affinity for cyt c with ATP (Figure S1) indicates that ATP competes with the lipid phosphate groups for cyt c binding, but the high peroxidase activity of CL-bound cyt c implies additional effects of ATP on the protein structure. Changes in the fluorescence of zinc-substituted cyt c with added ATP are in accord with this interpretation.16 ATP binding to cyt c decreases the protein stability,17 a factor that could contribute to the ease of cyt c unfolding and opening of the heme pocket, ultimately decreasing the population of C′ (Figure 3B). Even with the site S1 still interacting with CL liposomes, ATP may bind cyt c at other sites distant from the cyt c-CL binding interface (near Val20, Gly24 (S2) and Phe82 (S3)),2 (Figure 1) and destabilize the protein. Mutational and pH perturbations at or near these regions in cyt c have been implicated in an increased exposure of the heme.18,19
In summary, owing to its ionic nature, ATP disrupts cyt c-CL binding, causing some protein dissociation from liposomes. However, compared to the simple ionic effects of NaCl, ATP-induced changes in the protein conformational ensemble are distinct and suggest binding interactions that enable the CL-bound cyt c species to retain high peroxidase activity. Similar to the experimental conditions herein, the millimolar concentrations of ATP in mitochondria may play a role in modulating the CL-bound cyt c conformations and actually boosting cyt c apoptotic activity toward CL oxidation. Considering the strong interest in CL-bound cyt c, this study provides a critical platform for future analyses of structural features and the design of conformational regulators of this elusive protein ensemble.
Supplementary Material
Acknowledgments
Funding Sources: This work was supported by NIH RO1-GM098502 (E.V.P.). Funds to purchase the Nikon A1RSi Confocal Workstation were provided by NSF award DBI-1039423.
Footnotes
Notes: The authors declare no competing financial interests.
Supporting Information: Materials and methods; unfolding parameters for Dns variants; assays for cyt c-CL binding; fluorescence spectra, TR-FRET and distributions P(k) and P(r) for Dns variants; effects of different additives on population of E species in Dns92 cyt c; representative time course curves from peroxidase activity assays; FCS curves of NBD92 and Alx99 cyt c can be found free of charge at http://pubs.acs.org.
References
- 1.Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H. Proc Natl Acad of Sci USA. 2009;106:15651–6. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Forman HJ, Azzi A. FASEB. 1997;11:374–375. doi: 10.1096/fasebj.11.5.9141504. [DOI] [PubMed] [Google Scholar]
- 2.Patriarca A, Eliseo T, Sinibaldi F, Piro MC, Melis R, Paci M, Cicero DO, Polticelli F, Santucci R, Fiorucci L. Biochemistry. 2009;48:3279–3287. doi: 10.1021/bi801837e. [DOI] [PubMed] [Google Scholar]
- 3.Craig DB, Wallace CJ. Biochemistry. 1995;34:2686–93. doi: 10.1021/bi00008a036. [DOI] [PubMed] [Google Scholar]
- 4.Pletneva EV, Gray HB, Winkler JR. J Am Chem Soc. 2005;127:15370–15371. doi: 10.1021/ja0555318. [DOI] [PubMed] [Google Scholar]; Jordan T, Eads JC, Spiro TG. Prot Sci. 1995;4:716–728. doi: 10.1002/pro.5560040411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahluwalia U, Nayeem SM, Deep S. Eur Biophys J. 2011;40:259–71. doi: 10.1007/s00249-010-0643-6. [DOI] [PubMed] [Google Scholar]; Sinibaldi F, Mei G, Polticelli F, Piro MC, Howes BD, Smulevich G, Santucci R, Ascoli F, Fiorucci L. Prot Sci. 2005;14:1049–58. doi: 10.1110/ps.041069405. [DOI] [PMC free article] [PubMed] [Google Scholar]; Goto Y, Okamura N, Aimoto S. J Biochem. 1991;109:746–750. doi: 10.1093/oxfordjournals.jbchem.a123451. [DOI] [PubMed] [Google Scholar]
- 6.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Nat Chem Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 7.Sinibaldi F, Droghetti E, Polticelli F, Piro MC, Di Pierro D, Ferri T, Smulevich G, Santucci R. J Inorg Biochem. 2011;105:1365–1372. doi: 10.1016/j.jinorgbio.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 8.Beales PA, Bergstrom CL, Geerts N, Groves JT, Vanderlick TK. Langmuir. 2011;27:6107–6115. doi: 10.1021/la104924c. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bergstrom CL, Beales PA, Lv Y, Vanderlick KT, Groves JT. Proc Natl Acad of Sci USA. 2012 in press. [Google Scholar]
- 9.Brown LR, Wüthrich K. Biochim Biophys Acta. 1977;468:389–410. doi: 10.1016/0005-2736(77)90290-5. [DOI] [PubMed] [Google Scholar]; Salamon Z, Tollin G. Biophys J. 1996;71:848–857. doi: 10.1016/S0006-3495(96)79286-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanske J, Toffey JR, Morenz AM, Bonilla AJ, Schiavoni KH, Pletneva EV. Proc Natl Acad Sci USA. 2012;109:125–130. doi: 10.1073/pnas.1112312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rytömaa M, Kinnunen PK. J Biol Chem. 1994;269:1770–4. [PubMed] [Google Scholar]; Kostrzewa A, Pali T, Froncisz W, Marsh D. Biochemistry. 2000;39:6066–6074. doi: 10.1021/bi992559l. [DOI] [PubMed] [Google Scholar]
- 12.Hong Y, Muenzner J, Grimm SK, Pletneva EV. J Am Chem Soc. 2012;134:18713–18723. doi: 10.1021/ja307426k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maity H, Maity M, Englander SW. J Mol Biol. 2004;343:223–233. doi: 10.1016/j.jmb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Haldar S, Mitra S, Chattopadhyay K. J Biol Chem. 2010;285:25314–25323. doi: 10.1074/jbc.M110.116673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinibaldi F, Fiorucci L, Patriarca A, Lauceri R, Ferri T, Coletta M, Santucci R. Biochemistry. 2008;47:6928–6935. doi: 10.1021/bi800048v. [DOI] [PubMed] [Google Scholar]
- 16.Tuominen EK, Zhu K, Wallace CJ, Clark-Lewis I, Craig DB, Rytomaa M, Kinnunen PK. J Biol Chem. 2001;276:19356–19362. doi: 10.1074/jbc.M100853200. [DOI] [PubMed] [Google Scholar]
- 17.Antalík M, Bágel'ová J. Gen Physiol Biophys. 1995;14:19–37. [PubMed] [Google Scholar]
- 18.Feinberg BA, Liu X, Ryan MD, Schejter A, Zhang C, Margoliash E. Biochemistry. 1998;13091:13091–13101. doi: 10.1021/bi981037n. [DOI] [PubMed] [Google Scholar]; Rafferty SP, Pearce LL, Barker PD, Guillemette JG, Kay CM, Smith M, Mauk AG. Biochemistry. 1990;29:9365–9369. doi: 10.1021/bi00492a009. [DOI] [PubMed] [Google Scholar]
- 19.Balakrishnan G, Hu Y, Spiro TG. J Am Chem Soc. 2012;134:19061–19069. doi: 10.1021/ja307100a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.