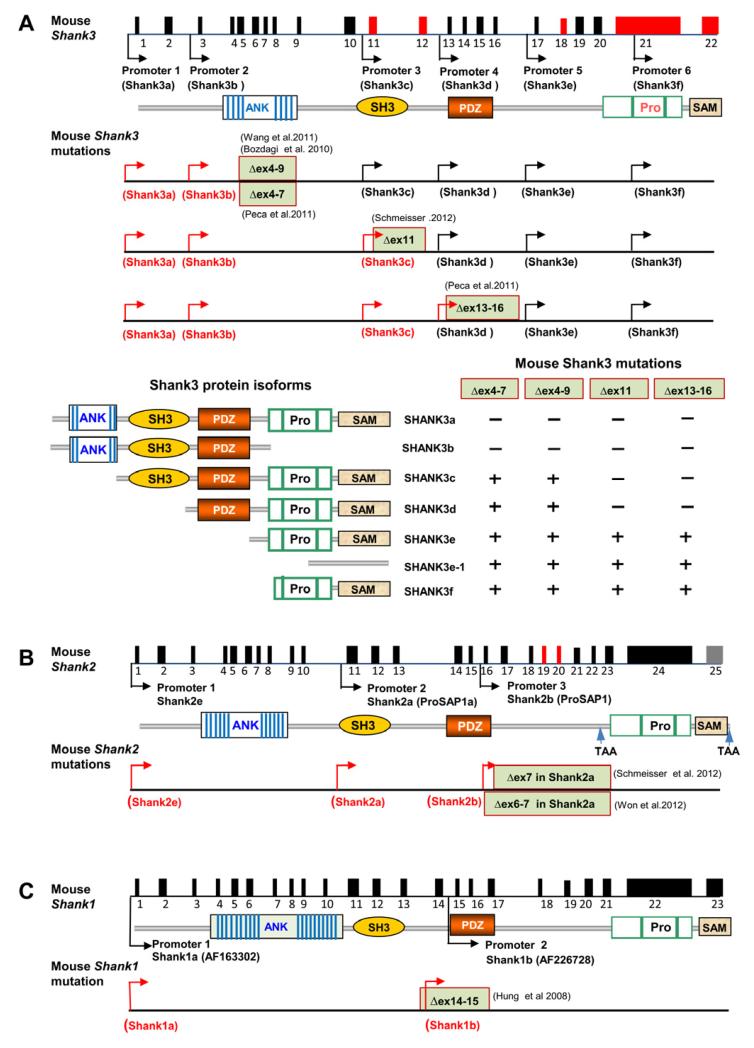
Figure 3.
Targeted Mutations in Shank Genes in Mice
(A) Schematic of the mouse Shank3 gene structure deduced from cDNA AB230103 deposited in GenBank. The promoters are shown by arrows and alternatively spliced exons are indicated in red. The positions of targeted mutations in five different lines of Shank3 mutant mice are shown. The transcripts from promoters upstream of deleted exons are predicted to be truncated or disrupted (red arrows) and the transcripts from promoter downstream of deleted exons are predicted to be intact in each mutant line of mice (black arrows). Bottom panels depict predicted isoform-specific expression of Shank3 mRNA and proteins in Shank3 mutant mice. The “−” indicates that the isoform is disrupted and “+” indicates the isoform remains intact. The full complement of Shank3 mRNA and protein isoforms that derive from combinations of alternative promoters and mRNA splicing remains unknown. Therefore, the pattern of isoform-specific expression and disruption by specific mutations is likely more complex than indicated.
(B) Schematic of mouse Shank2 gene structure and mouse mutations of Shank2. The sequence of Shank2 full length mRNA is not available and the intron-exon structure is deduced from a longest rat Shank2 cDNA deposited in GenBank (NM_201350). Exons in red are alternatively spliced. The human isoforms of SHANK2E, SHANK2A, and SHANK2B are conserved in mice (Shank2e, Shank2a, and Shank2b) but the status of SHANK2C is unknown. The Δex7 and Δex6-7 mutations (exon numbering based on the sequences of Shank2a/ProSAP1A cDNAs NM_001113373 or AB099695) described in the Shank2 mutant mice are shown (Schmeisser et al., 2012; Won et al., 2012). Exons 6 and 7 in Shank2a are deduced to correspond to exons 16 and 17 in the full length Shank2 gene structure diagram. The Shank2 Δex7 or Δex6-7 is predicted to cause truncation of all transcripts of Shank2 from the promoters upstream of exons 6-7 (red arrows).
(C). Schematic of mouse Shank1 gene structure and mouse mutation of Shank1. The deletion of exons 14-5 is predicted to disrupt all isoforms of Shank1 (Shank1a and 1b in red arrows) (Hung et al., 2008). In (A)-(C), genomic distance and exons are not drawn to scale.