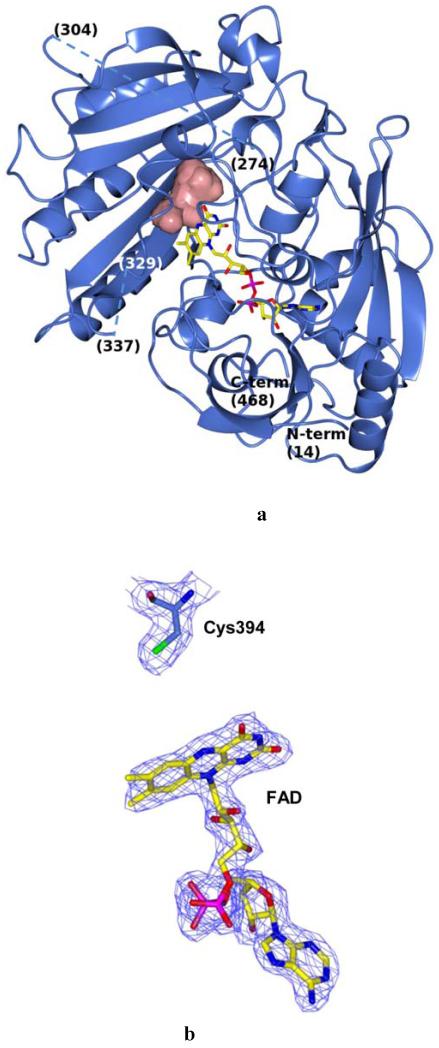
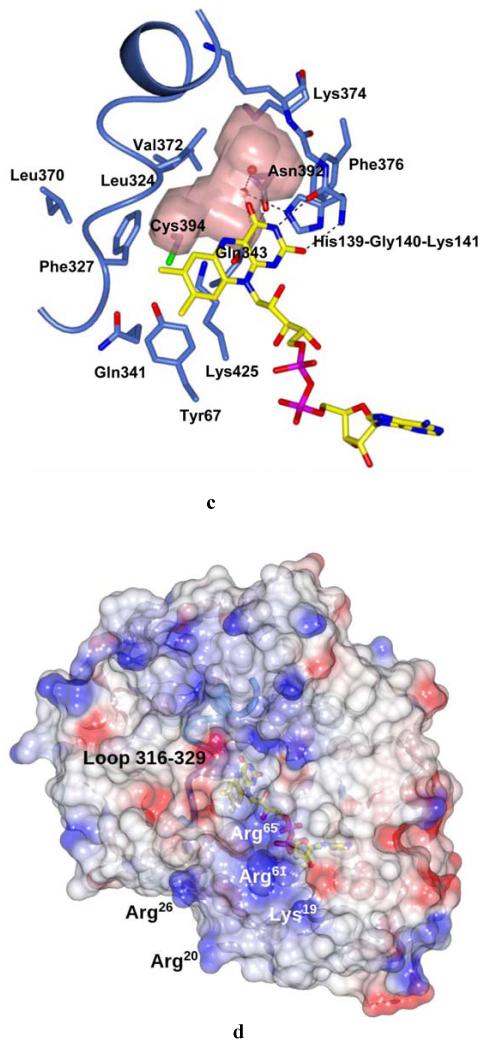
Fig. 2.
Crystal structure of M. smegmatis DprE1. (A) Light blue ribbon diagram of the DprE1 overall structure with labeled N- and C-termini. Disordered regions are indicated by dashed lines (numbers in parentheses correspond to the residues that flank the disordered parts). The FAD cofactor is represented in yellow with nitrogen, oxygen, and phosphorous atoms colored in blue, red, and magenta, respectively. The active site cavity is shown as pink surface. (B) Refined 2mFo-DFc electron density map (contoured at 1.2σ) for the FAD cofactor and the Cys394 known to be the target of covalent modification by BTZ043. For clarity, electron density contours further than 3Å from atoms in the figure have been omitted. Orientation and color code are as in Fig. 2A, except for protein carbon atoms, which are in light blue; sulfur atom is in green. (C) Close-up view of the DprE1 active site. Molecule orientation and color code are as in Fig. 2B.Water molecules are drawn as red spheres. Black dashed lines indicate H-bonds. The loop formed by residues 316-329 is shown, which shields the active site cavity from outside. (D) Electrostatic surface potential of DprE1 showing positively- and negatively-charged areas in blue and red, respectively. The FAD cofactor is visible in semi-transparency (color code as in Fig. 2B). Loop 316-329 is also visible in semi-transparency.