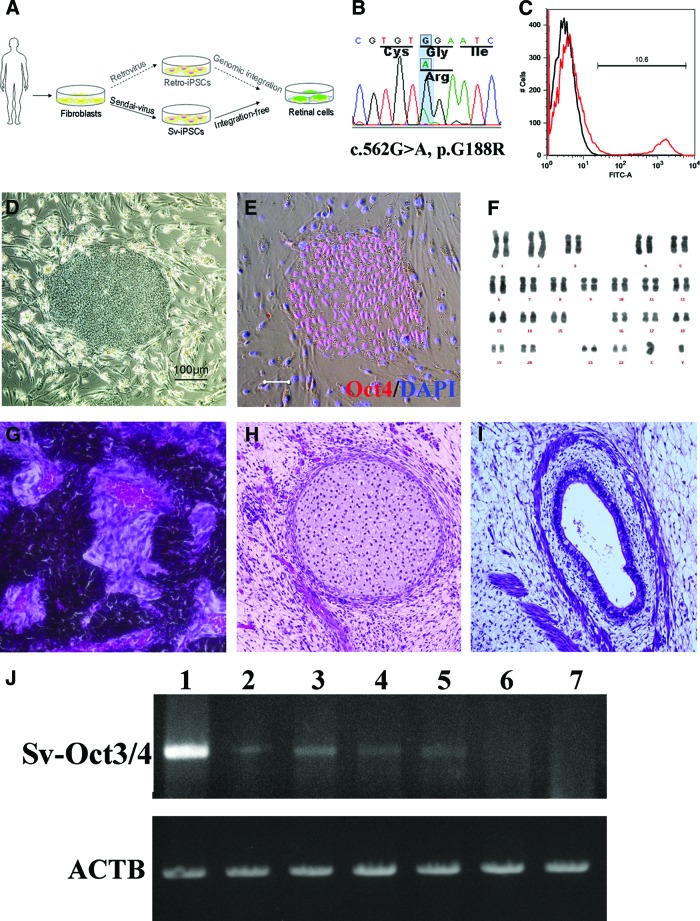
Figure 1.
Generation of patient-specific iPSCs. (A): Schematic diagram of this study. Compared with patient-iPSCs generated by the retroviral method, nonintegrative Sendai virus (SeV)-iPSCs from the same retinitis pigmentosa patient with a rhodopsin (RHO) mutation are more appropriate for disease modeling. (B): Sequence chromatogram of the RHO gene G188R mutation identified in patient-derived fibroblasts. (C): Representative result of SeV infection efficiency. The SeV infection efficiency was tested using SeV18+ green fluorescent protein (GFP)/TSΔF. Six days after infection of fibroblast cells, 10.6% of cells were positive for GFP (red histogram). Black histogram indicates a negative control. (D): The colony morphology of the patient-specific iPSCs. (E): Specific expression of the pluripotency marker Oct3/4 in the patient-derived iPSCs. Scale bar = 20 μm. (F): The results of karyotype analysis. (G–I): Histology of three germ layer derivatives in a teratoma formed by patient-derived iPSCs. (J): Ectopic expression of the Sendai viral Oct3/4 gene in the early passage iPSCs. Lane 1, positive control; lanes 2 and 3, 2nd passage iPSC lines of Sev3 and Sev9; lanes 4 and 5, 3rd passage Sev3- and Sev9-iPSC lines; lane 6, 10th passage Sev9-iPSC; lane 7, differentiated (day 40) Sev9-iPSCs. Abbreviations: ACTB, housekeeping β-actin gene; FITC, fluorescein isothiocyanate; iPSCs, induced pluripotent stem cells; Sv, Sendai virus; Sv-Oct3/4, Sendai viral Oct3/4 gene.