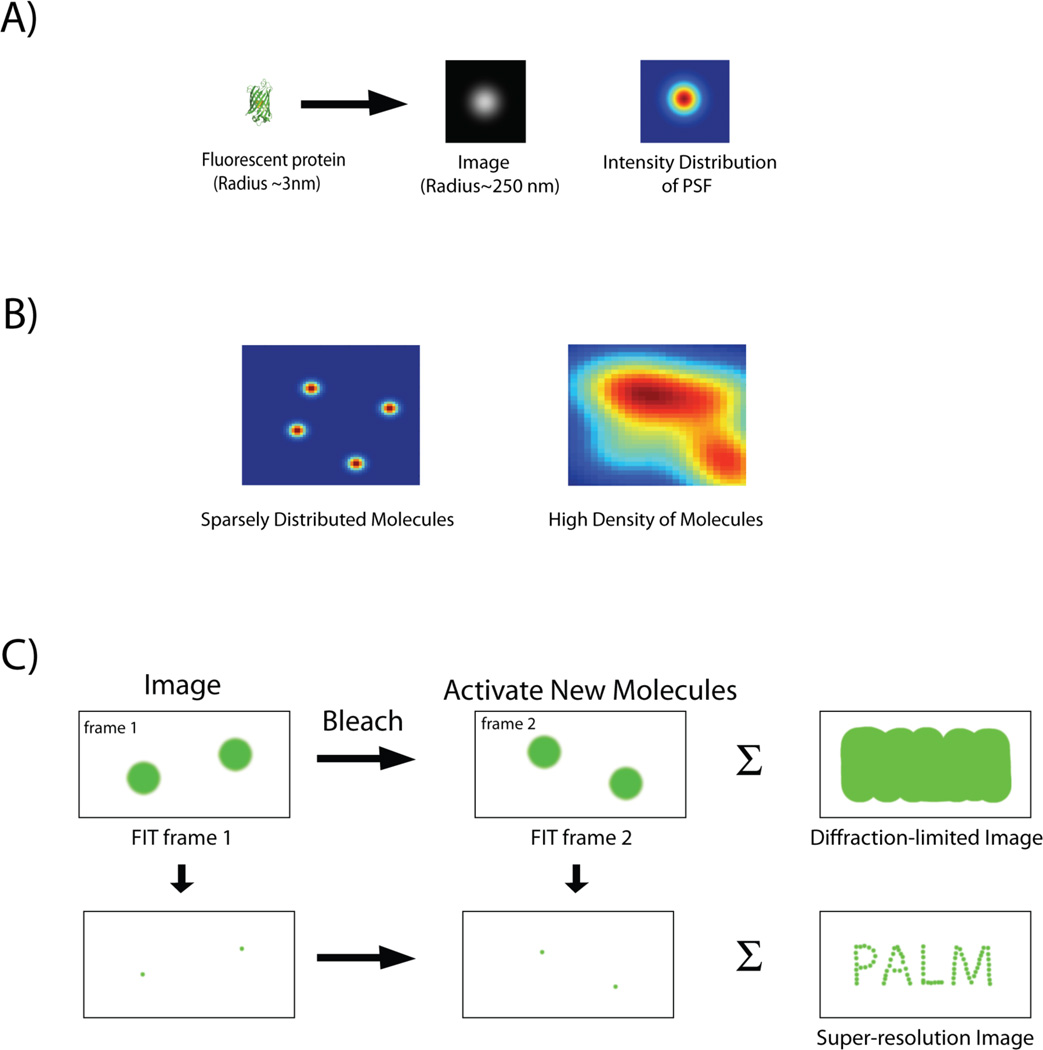
Figure 1.
A: Image of a single fluorescent protein is convoluted by the point spread function (PSF) of the imaging system and appears as an extended blob. B: Intensity distribution of fluorescent proteins expressed at different densities. Images of single fluorescent proteins that are spatially separated by distances greater then the width of the PSF can be individually fit and localized with high precision. In contrast, PSFs of closely spaced molecules merge (high density of molecules), and individual molecules can no longer be distinguished. C: Imaging protocol for PALM. A sparse subset of photoactivable proteins is activated, imaged and their localization is determined with subdiffraction precision by a 2-D Gaussian fit. Following photobleaching of the activated proteins, a new set of molecules is activated, and the cycle is repeated. Finally the individual localizations are combined together to construct a super-resolution image, which depicts structural details unobservable by conventional, diffraction-limited imaging. The summation of the diffraction-limited spots, in contrast, gives back an image, which is equivalent to one captured by conventional optical microscope.