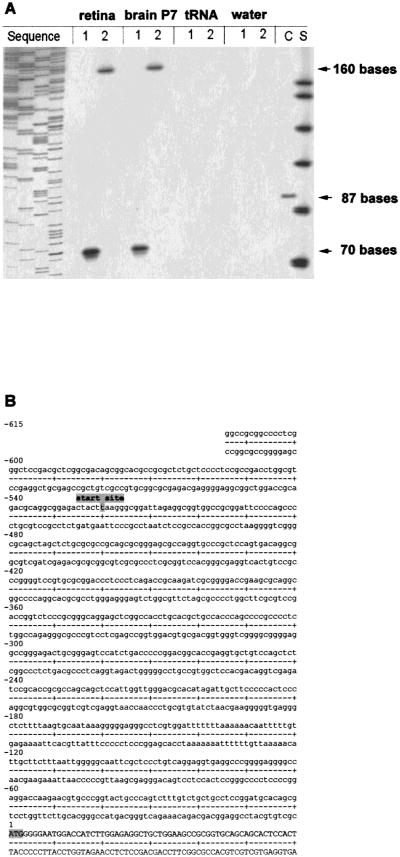
Fig. 3.
(A) Primer extension analysis of mouse Cx36 transcript in retina and brain and (B) Nucleotide sequence of the 5′ untranslated region of mouse Cx36. (A) Mouse retina and mouse brain RNA were annealed to Cx36 specific primers Pex1 (1) and Pex2 (2). The primers were extended using reverse transcriptase and the products were electrophoretically separated and visualized by autoradiography. Arrows indicate the size of different extension products, i.e., 70 bases for primer Pex1 and 160 bases for primer Pex2. No products were seen in control reactions using tRNA or water. Based on the length of both extension products, the transcription start point was mapped to position −520 upstream of the translation start point. The sizes of the extension products were determined with the standard S (ΦX174/HinfI DNA marker [Promega], 151, 140, 118, 100, 87, 82 and 66 bases). The sequencing reaction on the left side helped to determine the product sizes more precisely. In the control reaction (C) a synthetic mRNA coding for the kanamycin resistence gene (Promega) was extended to 87 bases. (B) The transcriptional start point is indicated at position −520. The translational start codon is at position +1. Cx36 exon I comprizes 591 bp, of which 520 bases are untranslated and 71 bases are part of the coding region.