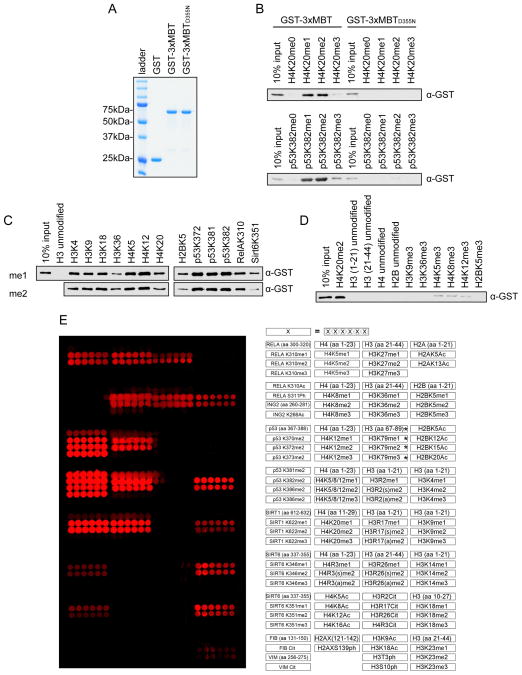
Figure 1.
3xMBT recognizes methylated lysine with broad sequence specificity. (A) Coomassie stain of glutathione S-transferase (GST) alone, GST-tagged 3xMBT, and GST-tagged 3xMBTD355N. (B) 3xMBT, but not methyllysine-binding mutant 3xMBTD355N, binds to mono- and di-methylated H4K20- and p53K382-containing peptides. Western blot analysis of peptide pull-downs with GST-3xMBT and GST-3xMBTD355N and the indicated biotinylated peptides. H4 peptides: amino acids 1–23. p53 peptides: amino acids 367–388. (C) 3xMBT coprecipitates with a panel of biotinylated mono- and di-methylated peptides with varying sequences. Peptides are labeled by their methylated residues; all peptides are roughly 20 amino acids in length, with only the indicated residues methylated. (D) 3xMBT does not bind to a series of non-methylated peptides. Pull-down experiments with the indicated peptides as in (C). (E) 3xMBT binds preferentially to diverse mono- and di-methylated peptides. Microarrays spotted with over 100 distinct histone and non-histone peptides as indicated in the array key on the right were probed with GST-3xMBT. Red spots indicate positive binding. 3xMBT does not bind unmodified peptides, tri-methylated peptides, or peptides bearing other post-translational modifications. *H3K79 peptides have low solubility and therefore are not transferred efficiently to the array surface. See also Figure S1.