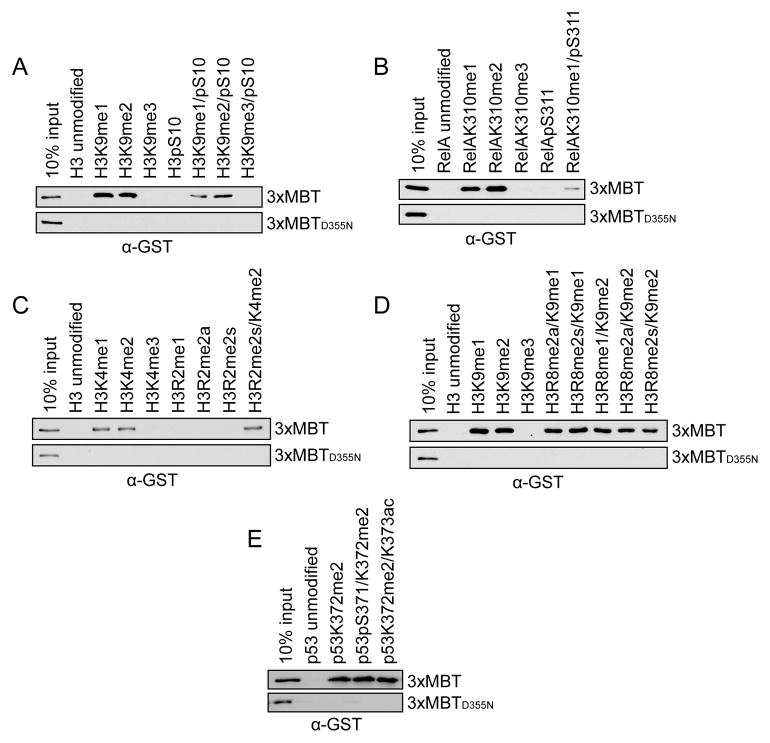
Figure 2.
3xMBT recognizes mono- and di-methyl lysine in the presence of secondary posttranslational modifications. (A–E) Western blot analysis of peptide pull-down assays with GST-3xMBT and GST-3xMBTD355N and the indicated biotinylated peptides. All modified peptides have the same base sequence and length as the unmodified peptide control. (A) Phosphorylation of H3S10 modestly decreases binding of 3xMBT to mono- and di-methyl H3K9. (B) Phosphorylation of RelA at S311 decreases, but does not abolish, binding of 3xMBT to RelAK310me1. (C) Symmetric dimethylation of H3R2 does not block 3xMBT binding to H3K4me2-containing peptides. (D) Different states of methylation of H3R8 have little effect on binding of 3xMBT to H3K9me1- and H3K9me2-containing peptides. (E) 3xMBT binds p53K372me2 in the presence of adjacent serine phosphorylation or lysine acetylation. See also Figure S2.