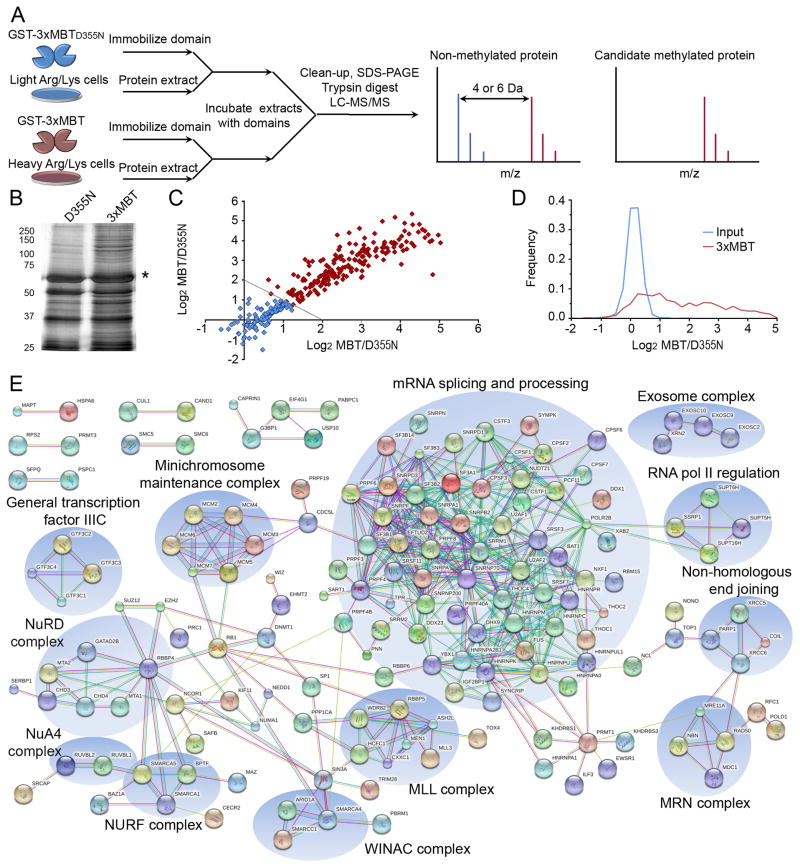
Figure 5.
Protein pull-down using 3xMBT reproducibly enriches for the methyl-lysine proteome. (A) Protocol schematic for proteome-wide capture and identification of candidate methylated proteins using 3xMBT. Peptides derived from specifically enriched proteins show a distinctive signal in the SILAC label associated with 3xMBT, while peptides from non-specifically bound proteins (equal binding to 3xMBT and 3xMBTD355N resins) have the same intensity in both SILAC conditions. (B) Abrogating the methyl-lysine binding site of 3xMBT greatly reduces the amount of protein captured from nuclear extract. Silver stain of total proteins present in GST-3xMBT and GST-3xMBTD355N pull-downs from 293T nuclear extracts. Asterisk indicates GST fusion band. (C) Specific proteins are reproducibly and quantitatively enriched by 3xMBT relative to 3xMBTD355N. Axes represent the quantitative ratio (log base 2) for 3xMBT over 3xMBTD355N in independent experiments. Each point represents a protein identified in both experiments, and enrichment in these experiments is correlated with R2=0.86. (D) Most proteins identified following enrichment by 3xMBT (approximately 60%) are specifically enriched by the methyl-lysine binding domain. Probability density plots show 3xMBT enrichment for all 544 identified proteins identified in pull-down experiments compared to the SILAC ratios of proteins identified from input material. (E) Proteins enriched by 3xMBT tend to be functionally related based on protein interaction networks. The diagram shows all high-confidence protein-protein interactions from the STRING interaction database between proteins enriched to at least 2-fold by 3xMBT relative to 3xMBTD355N. See also Tables S2 and S3.