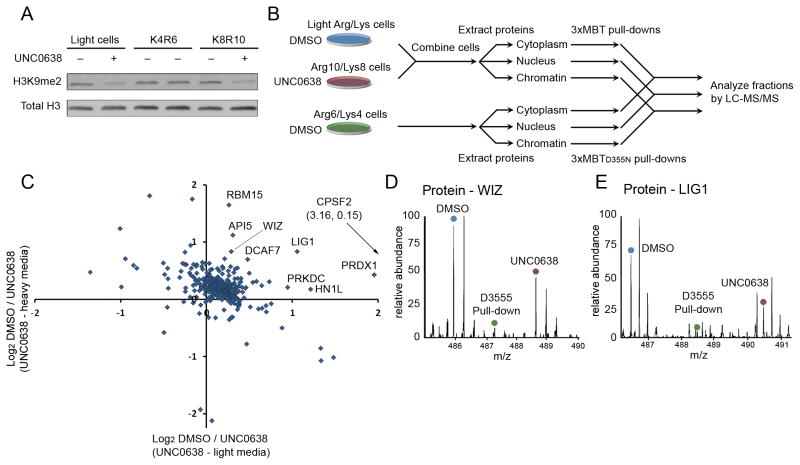
Figure 6.
Candidate physiologic substrates of G9a and GLP are identified by targeted inhibition and quantitative proteomic analysis. (A) Treatment with UNC0638 decreases global H3K9me2 levels. Western blot of 293T whole cell extracts ± UNC0638 treatment and probed with the indicated antibodies. (B) A schematic diagram of the proteomic experiment. Cells grown in “light” or “heavy” media were treated with DMSO or the G9a/GLP inhibitor UNC0638 and combined prior to extracting proteins from cytoplasmic, nuclear and chromatin fractions. Cell prepared in “medium” media were treated with DMSO, processed and fractionated in parallel. Light/heavy extracts were subjected to pull-down by 3xMBT, while “medium” extracts were subjected to pull-down by 3xMBTD355N. Bound proteins from each fraction were combined and analyzed by LC-MS/MS. (C) Identification of dozens of candidate in-cell substrates of G9a/GLP. Many proteins show decreased association with 3xMBT following 24 hour treatment of 293T cells with UNC0638 relative to DMSO treatment. Only proteins that were also enriched by 3xMBT relative to 3xMBTD355N are shown. Axes show log2 ratios for levels from vehicle control over treated cells, and each axis represents an independent biological replicate. (D) MS1 spectrum showing that the amount of WIZ, a known G9a substrate, captured by 3xMBT from cell lysate is reduced following treatment with UNC0638. The source of the relevant peak is indicated. (E) MS1 spectrum showing reduction in LIG1 captured by 3xMBT following treatment with UNC0638 as in (D). See also Figure S4 and Table S4.