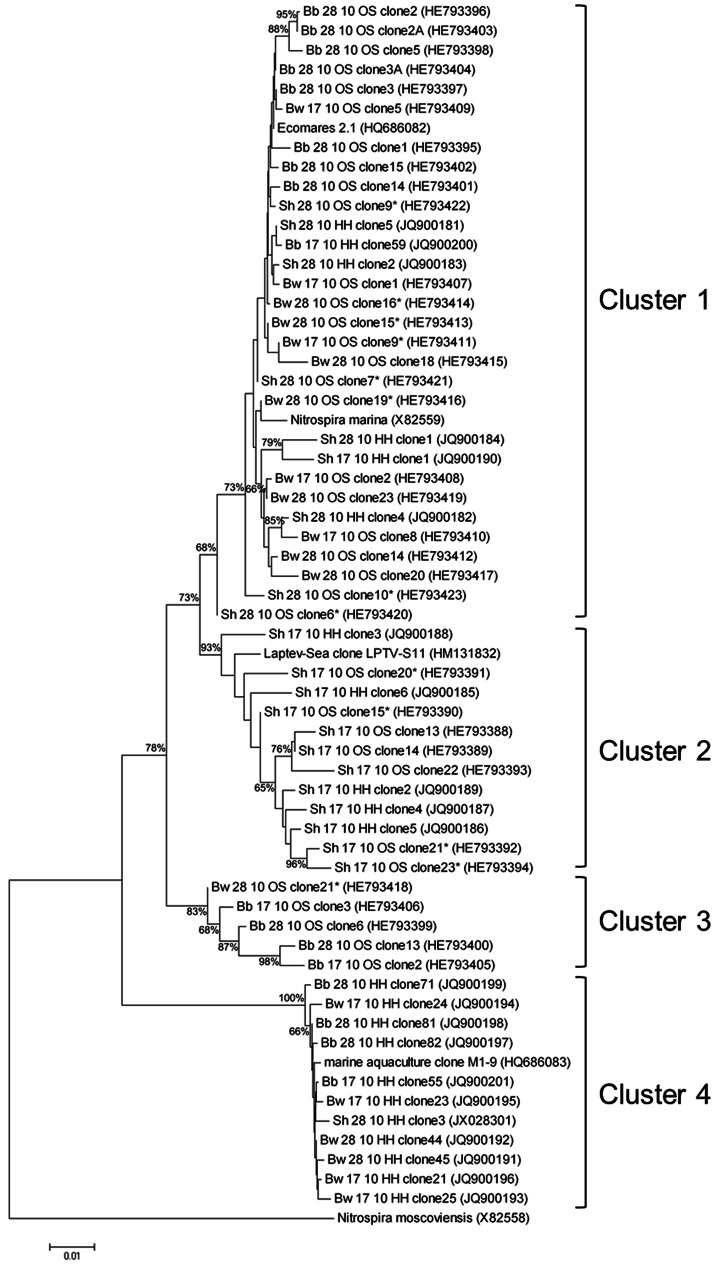
Figure 3. Phylogenetic tree based on 16S rRNA gene sequences.
The tree was constructed by neighbor-joining algorithm showing four different clusters (1–4). Nodes ≥65% supported by bootstrap values (based on 1000 iterations). The tree shows sequences of the marine Nitrospira sublineage IV with clones of the brackish RAS biofilters. The sequence of Nitrospira moscoviensis from sublineage II was defined as outgroup. The sequences obtained from analyzed biofilters were named after their origin (Bw: barramundi white biocarrier; Bb: barramundi black biocarrier; Sh: shrimp biocarrier), the incubation temperature (17 or 28°C), as well as the appendix OS (acc. nos. HE793388 – HE793423) and HH (acc. nos. JQ900181-JQ900201; JX028301) for different cloning approaches. Sequences ≤800 bp was marked with “*”. Scale bar = 1% sequence divergence.