Figure 2. Effect of mutation frequency on DH and DRF-identification accuracy.
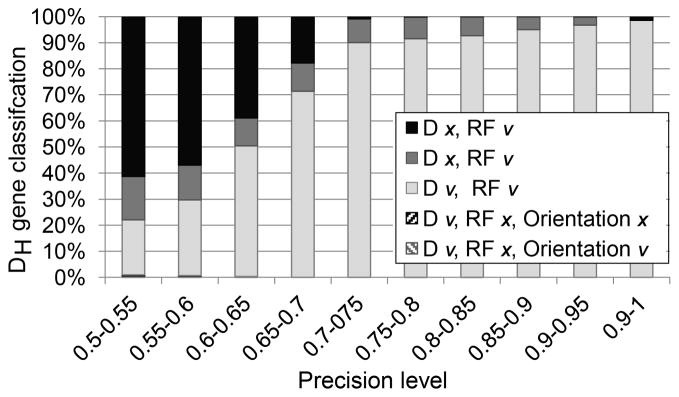
The X-axis is the fraction of nucleotides in the sampled sequence that overlaps with the full DH germline sequence. The Y-axis is the fraction of properly classified DH genes in simulated rearrangements. The errors were divided into multiple categories including errors in DH identification or in the DRF classification or both. A detailed analysis of the effect of the DH gene on the precision is provided in Supplemental Fig. 1. One can clearly see that as soon as the overlap is higher than 0.7, the fraction of sequences with a reading frame error is less than 1 % and the DH identification error is less than 10 %. Most of those occur in highly similar DH genes. Reading frames with a negative number are inverted reading frames. Those are observed only in very short DH genes.
