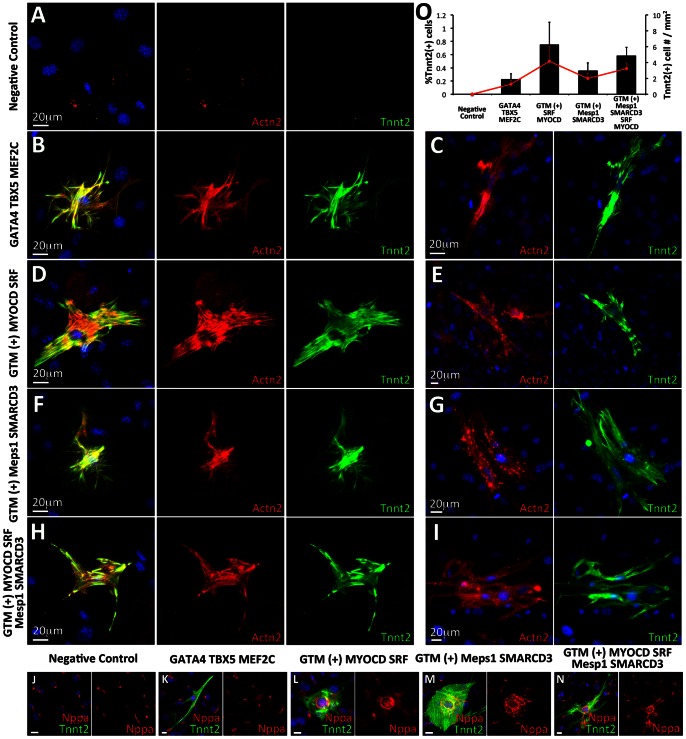
Figure 5. Reprogrammed MEFs express cardiac specific proteins and organize them in a cross-striated manner.
A–I. Induction of TF overexpression for 7 days in MEFs transduced with only FUW.M2rtTA or the four listed combinations of TF modules. The reprogrammed cells were cultured on gelatin-coated plastic in low serum growth medium. Using double-antibody immunofluorescence analysis (Actn2/Red, Tnnt2/Green) we detected cells expressing both cardiac proteins and organizing them in a cross-striated manner resembling cardiomyocytes (B, D, F, H). We detected significantly more double-positive cells in MEFs transduced with G4T5MCMDSF. For each of the transcriptional module combinations we also detected double-positive cells without any obvious cross-striated cytoskeletal organization (C, E, G, I). No Actn2, or Tnnt2 cross-striated expression was detected in the negative control cells. J–N. The Tnnt2 expressing cells also stained positive for the atrial protein marker Nppa. O. Quantification of the fraction of cells staining positive for the Tnnt2 cardiac protein (low serum growth medium) as compared to the total number of cells (black columns) and measurement of the fraction of Tnnt2-expressing cells per square millimeter (red line). Results are based on biological triplicates. Error bars represent calculated standard deviation. All four cell groups had a significant increase in the number of Tnnt2(+) cells as compared to the negative control group (P<0.01). Cells transduced with either G4T5MCMDSF (P<0.01), G4T5MCMDSFM1S3 (P<0.05), G4T5MCMDSFM1S3 (P<0.01) also had a significant increase in the number of Tnnt2(+) cells as compared to cells transduced with G4T5MC.