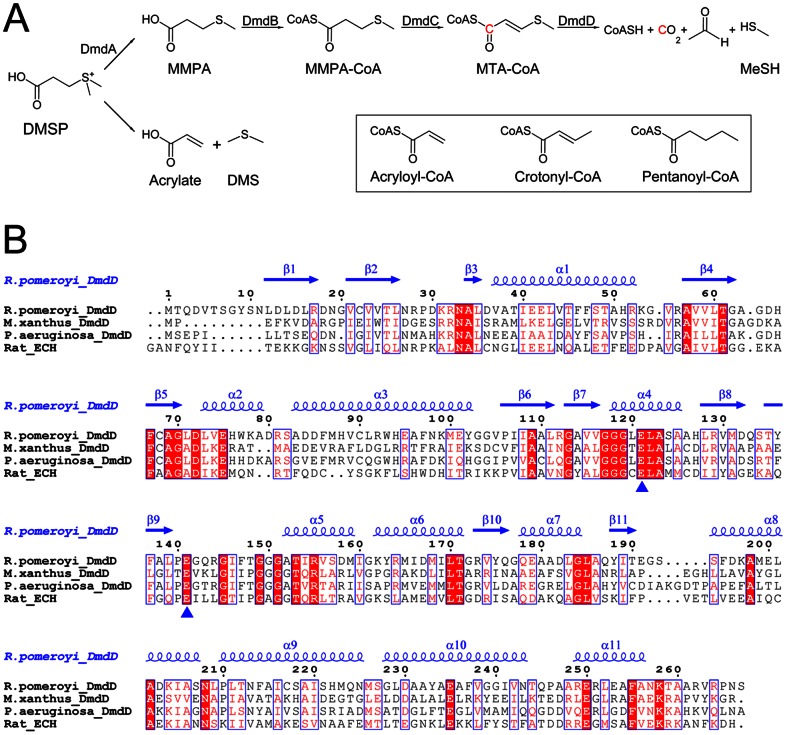
Figure 1. Sequence conservation of DmdD.
(A). The MeSH and DMS catabolic pathways of DMSP. The enzymes in the MeSH parthway are indicated. The chemical structures of acryloyl-CoA, crotonyl-CoA and pentanoyl-CoA are shown in the inset. (B). Sequence alignment of Ruegeria pomeroyi DmdD, Myxococcus xanthus DmdD, Pseudomonas aeruginosa DmdD, and rat liver enoyl-CoA hydratase (ECH). The secondary structure elements in the structure of R. pomeroyi DmdD are labeled. The two catalytic Glu residues are indicated with the blue triangles. The residue numbers refer to R. pomeroyi DmdD.