Figure 1.
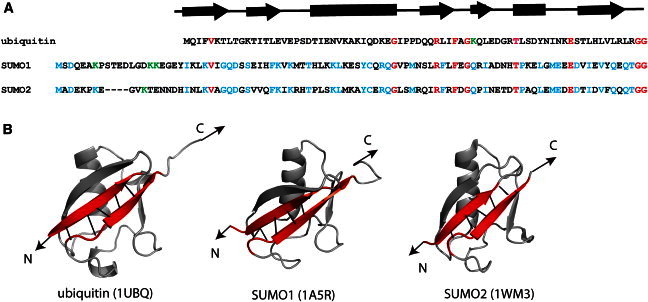
(A) The amino acid sequences of ubiquitin, SUMO1, and SUMO2. The residues that are identical in all three proteins are highlighted in red. In SUMO1 and SUMO2 there exists a long structureless sequence (∼20 residues) at N-terminus, which is absent in ubiquitin. The sequence identity between SUMO1/ubiquitin, SUMO2/ubiquitin pairs is ∼16%. The sequence identity between the SUMO1/SUMO2 pair is high (∼50%) and the residues identical in both of them are highlighted in blue. The C-terminal diglycine sequence is preserved in all of them. Ubiquitin forms Lys-48-C-linked polyubiquitin, whereas SUMO2 forms Lys-11-C-linked polymer chains. SUMO1 forms polymer chains in vitro through Lys-7, Lys-16, and Lys-17, which are highlighted in green. The secondary structural elements of ubiquitin are indicated directly above the sequence. (B) Structures of SUMO1, SUMO2, and ubiquitin. PDB IDs are given in parenthesis. The N-terminus structureless regions in SUMO1 and SUMO2 are not shown. In all the structures, the terminal β-strands (shown in red) are parallel to each other and connected by five H-bonds between the backbones (black lines). The arrows indicate the pulling direction used in the mechanical unfolding experiments.
