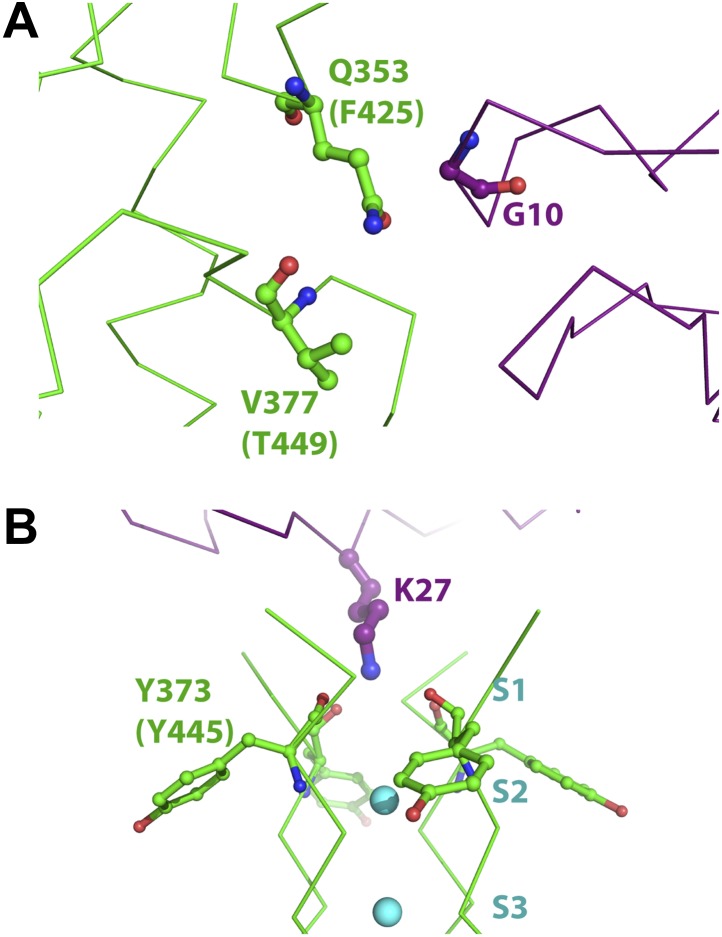
Figure 10. Mutant cycle data from Ranganathan et al. (1996) mapped onto the structure of paddle chimera–CTX complex.
The toxin molecule shown in purple is AgTx2, which was superimposed onto CTX in the structure of the paddle chimera–CTX complex using the NMR structure of AgTx2 (PDB ID 1AGT; Krezel et al., 1995). Guidelines in Krezel et al. (1995) were used for the superposition. (A) Close-up view of AgTx2 showing Glycine 10 in ball and stick rendition and the rest of the toxin in α carbon trace. The channel subunit that is most proximal to Gly10 is shown in green α carbon trace. Shown in ball and stick are residues Gln353 and Val377 in paddle chimera, that correspond to Phe425 and Thr449, respectively, in Shaker using a sequence alignment (Figure 2B). Ranganathan and MacKinnon reported that Gly10 is coupled to Phe425 and Thr449 in Shaker. (B) Close-up view of AgTx2 showing the side-chain of Lys27 in ball and stick rendition and the rest of the toxin in α carbon trace. The nearby regions of the selectivity filter from all four subunits of the channel are shown in green α carbon trace. Shown in ball and stick is Tyr373 in all four subunits of paddle chimera, which map onto Tyr445 in Shaker using a sequence alignment (Figure 2B). Ranganthan and MacKinnon reported Tyr445 to be coupled to Lys27, and this was dependent on the concentration of K+.