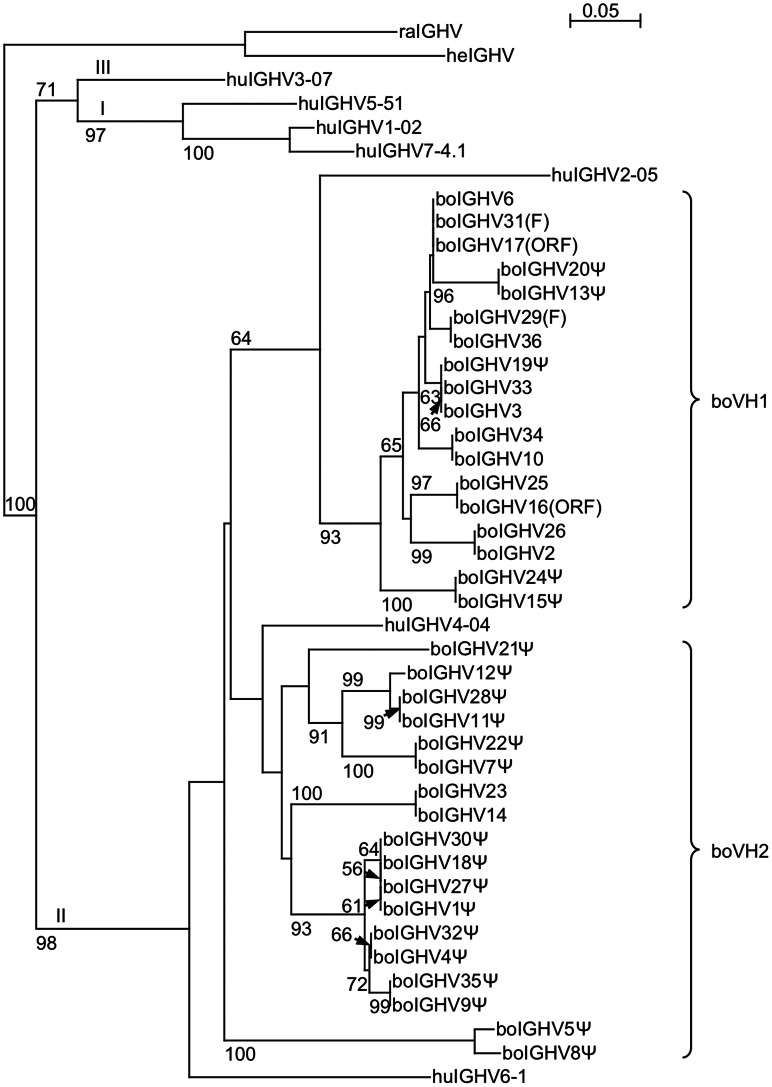
Figure 2. Neighbor-joining phylogenetic tree of the genomic bovine IGHV segments.
The complete sequences of the bovine IGHV segment (boIGHV), and one representative sequence of each human family (huIGHV1 to huIGHV7), were used for the comparison. The reliability of the tree was estimated using 1000 bootstrap replicates [43]. Numbers at each node are the percentage bootstrap value and are indicated only when greater than 50%. Arrows mark the respective node. The Roman numerals I, II, and III describe the clans [64]. Two clusters of bovine IGHV were visible and corresponded to two families. The bovine IGHV family 1 (boVH1) comprises all functional segments, whereas boVH2 consists only of pseudogenes. IGHV5Ψ and IGHV8Ψ present fragmented loci, which consist of only 77 bp. They share 70.1% sequence identity with IGHV1Ψ, IGHV18Ψ, IGHV27Ψ, and IGHV30Ψ. We would therefore propose to assign IGHV5Ψ and IGHV8Ψ to boVH2. Horned shark (heIGHV from accession number X13449) and little skate (raIGHV; X15124) represent the outgroup in this analysis, similar to that performed by Sitnikova and Su [44] and Almagro et al. [45]. The scale bar indicates the number of nucleotide substitutions per site.