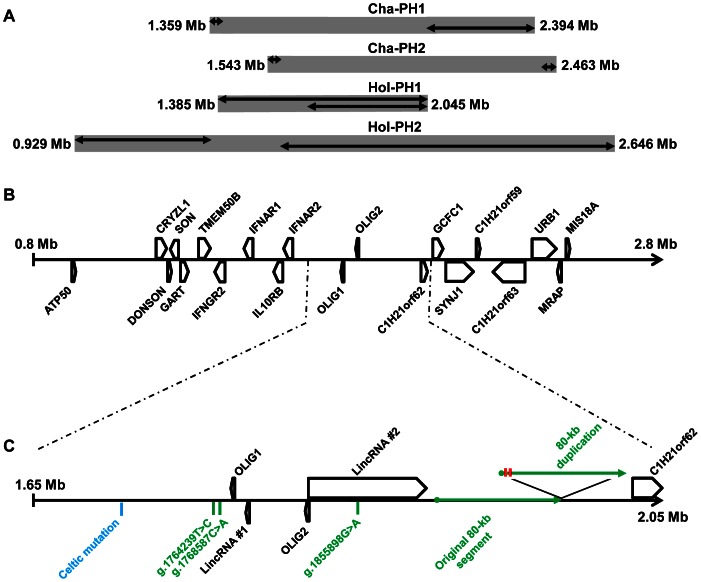
Figure 2. Accurate mapping of the Polled locus and identification of candidate causative mutations.
(A) Localization on BTA01 of the intervals (gray boxes) containing the Polled mutations associated with different haplotypes based on Illumina BovineSNP50 beadchip genotyping data. Upper and lower double arrows indicate the region in which the most informative recombination occurred at the left and right border of the interval, respectively. (B) Gene content of the Polled intervals. (C) Localization of the candidate mutations for the Celtic (blue) and Friesian (green) Polled alleles, and details of the coding and non-coding genes in their close vicinity. LincRNA: Long intergenic non-coding RNA. LincRNA#1 and 2 correspond, respectively, to EST sequences n° AW356369, AW357421, BF654718, BM105296, BM254775, BM254845 and BC122836, DT831326, DT837875, DY200702, DY169884, EH130782, EH138227, EV606908, EV693397 in Genbank. Positions on BTA01 are based on the UMD3.1 bovine genome assembly. Polymorphism g.1855898G>A is located within an intron of LincRNA#2. Red bars indicate the two sequence variations between the original and the duplicated 80-kb segments.