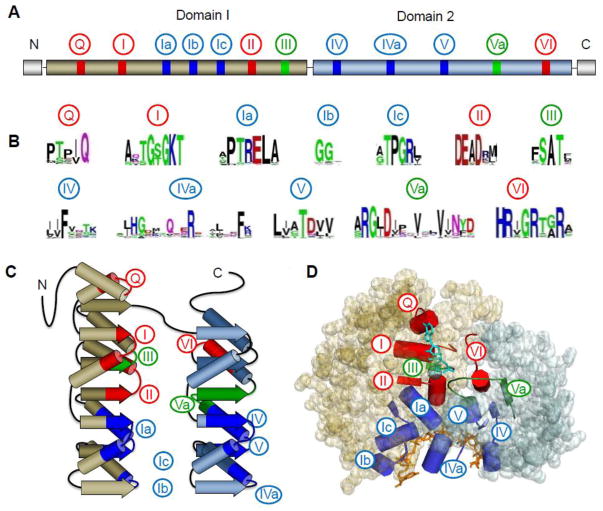
Figure 1. Architecture of the DEAD-box helicase core.
(A) Schematic of the primary structure of the DEAD-box helicase core. Domain 1 (N-terminal, tan) and 2 (C-terminal, blue) designate the two RecA-like helicase domains. Circled numbers indicate the approximate location of the characteristic sequence motifs, colored according to their primary biochemical function (red, ATP binding and hydrolysis; blue, RNA binding, green, coordination between RNA and ATP binding). (B) Sequence conservation in the characteristic sequence motifs of DEAD-box helicases. Amino acid conservation is represented by the height of letter. Coloring marks the chemical properties of a given amino acid: green and purple — polar, blue — basic, red— acidic, and black— hydrophobic (adapted from ref. [1]). (C) Schematic representation the topology of the two RecA-like helicase core domains. β-strands are indicated by arrows, α-helices by cylinders. Characteristic sequence motifs are colored as in panel A (adapted from ref [1]). (D) Position of the characteristic sequence motifs in the three-dimensional structure of the DEAD-box helicase Vasa [19]. The bound ATP analog and Mg2+ ion are colored teal; the nucleic acid is colored orange. Sequence motifs are colored as in panels A to C.