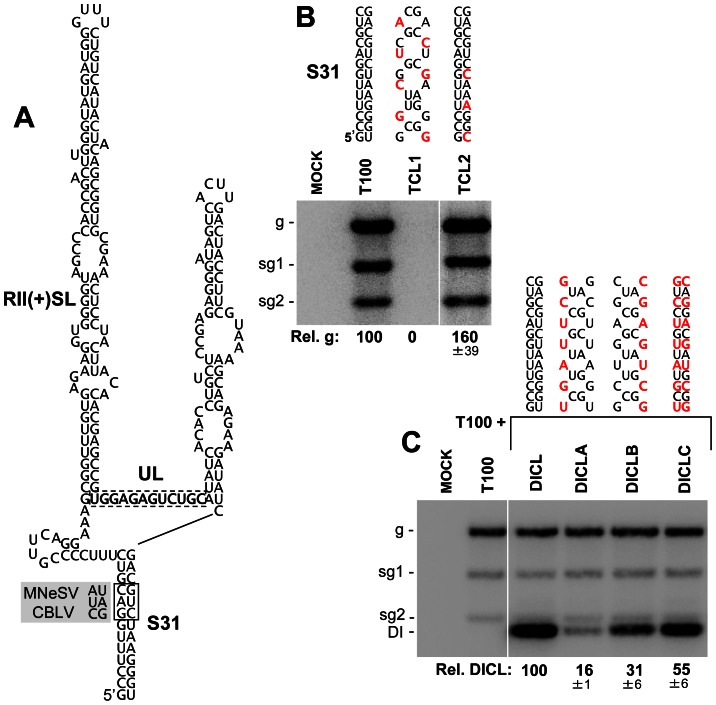
Figure 7. Mutational analysis of the predicted stem-31 structure.
(A) Predicted structure for S31. Base pairs in the structure that covary in MNeSV and CBLV are boxed by a solid line and the substituted nucleotides are shown in the shaded regions to the left. The UL is boxed by a dotted line and RII(+)SL, which corresponds to S33 in the genome model, is indicated. (B) Northern blot analysis of viral RNA accumulation in protoplasts transfected with TBSV genome mutants. The name and changes made in S31 in the genomic mutants are shown above each lane, with substitutions depicted in red. Relative TBSV genome accumulation is shown at the bottom. Values represent means with standard errors from three independent experiments. (C) Northern blot analysis of DI RNA accumulation in protoplasts co-transfected with wt TBSV genome and wt or mutant forms of DICL. The changes in S31 in the DICL mutants are indicated for each lane. Quantification of the DI RNAs is shown below the blot. Values represent means with standard errors from three independent experiments.