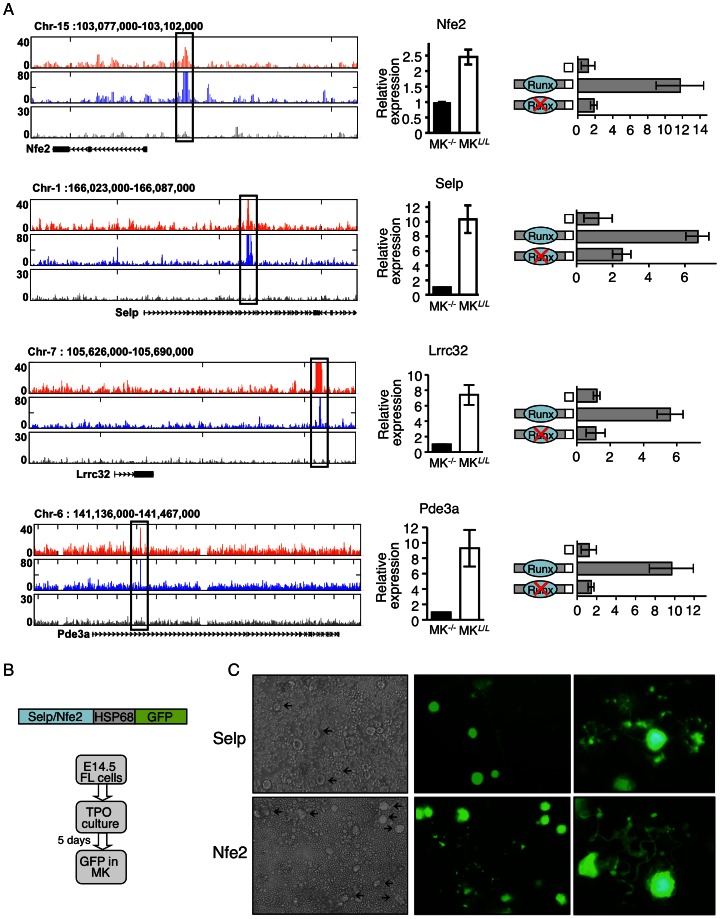
Figure 7. Validation of Runx1/p300 bound megakaryocytic enhancers.
(A) Transfection-mutagenesis validation of Runx1 dependent activation of Nfe2, Selp, Lrrc32 and Pde3a regulatory elements by transfection into megakaryocytic cell line Meg01. Tracks of Runx1 (orange), p300 (blue) and non-immune serum (gray) ChIP-seq traces in maturating FL-MK (left panels). The cloned Runx1/P300 bound regions are marked by quadrangles. RT-qPCR analysis recording expression of the genes in FL-MKRunx1−/− (MK−/−), or FL-MKRunx1L/L (MKL /L) (middle panels). The data represent means ± SD of two experiments performed in triplicates. Dual luciferase reporter assays in transfected Meg01 cells using PGL4.73 vector alone or with the indicated intact/mutated regulatory region. (B) Transgenic analysis of Selp and Nfe2 regulatory regions. Co-bound Runx1/p300 regulatory regions were cloned upstream of the HSP68 promoter-GFP constructs. FL cells of E14.5 PCR-GFP-positive transgenic embryos were cultured with TPO 50 ng ml−1 for 3–5 days before evaluation of GFP expression. (C) Light and fluorescence microscope images of 4-days cultured FL cells derived from E14.5 Selp (15/26 positive embryos) or Nfe2 (11/20 positive embryos) transgenic embryos. Images were produced using an Olympus IX71 microscope with 10×/0.3 (left and middle) and 40×/0.6 (right) objective lenses.