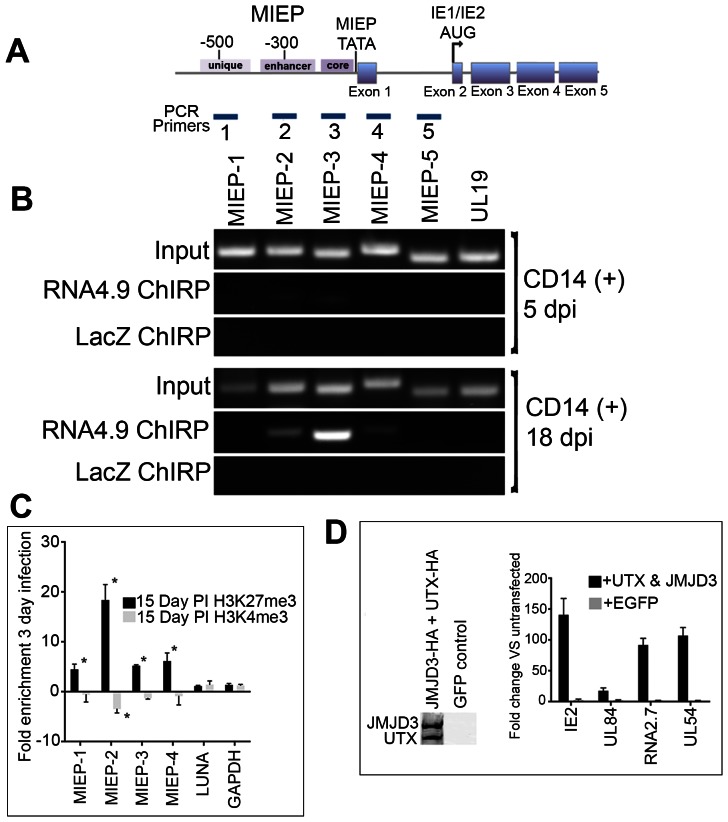
Figure 7. RNA4.9 physically interacts with the HCMV latent viral chromosome.
(A) Schematic of the HCMV MIE gene and promoter/enhancer region. Regions amplified by PCR primers are shown that are specific for the unique, enhancer, core promoter, exon 1 and the first intron of IE2. (B) ChIRP analysis of RNA4.9 binding to regions of the MIE promoter and gene locus. Infected CD14 (+) cells were harvested at 5 and 18 days post infection and ChIRP was performed. Control ChIRP was performed using biotinylated hybridization primers specific for LacZ. Amplification of the UL19 region was used as a control for ChIRP PCR amplification. (C) Increase in the enrichment of the repressive H3K27me3 mark at the MIEP during latency is consistent with the binding of RNA4.9. CD14 (+) cells infected with HCMV were harvested at 3-days post infection or during latency at 15 days post infection and ChIP assays were performed using antibodies specific for H3K27me3, H3K4me3 or IgG control antibody. Fold enrichment of H3K27me3 or H3K4me3 was calculated by IgG subtracted % input of each locus divided by the IgG subtracted % input of the control gene GAPDH. Each sample was evaluated in triplicate and the error bars are the SD of the mean. Data is shown as fold enrichment compared to tri methylation state at 3 days PI. Statistical analysis was done using multiple t-test, *P Value<0.001. (D) Increase in IE mRNA accumulation in cells transfected UTX and JMJD3 expression plasmids. Latently infected CD14 (+) cells were transfected with plasmids expressing UTX and JMJD3. Expression of UTX and JMJD3 was confirmed by Western blot, left panel. Graph shows the increase in fold accumulation of specific mRNAs. Error bars are the standard deviation of the average of three experiments.