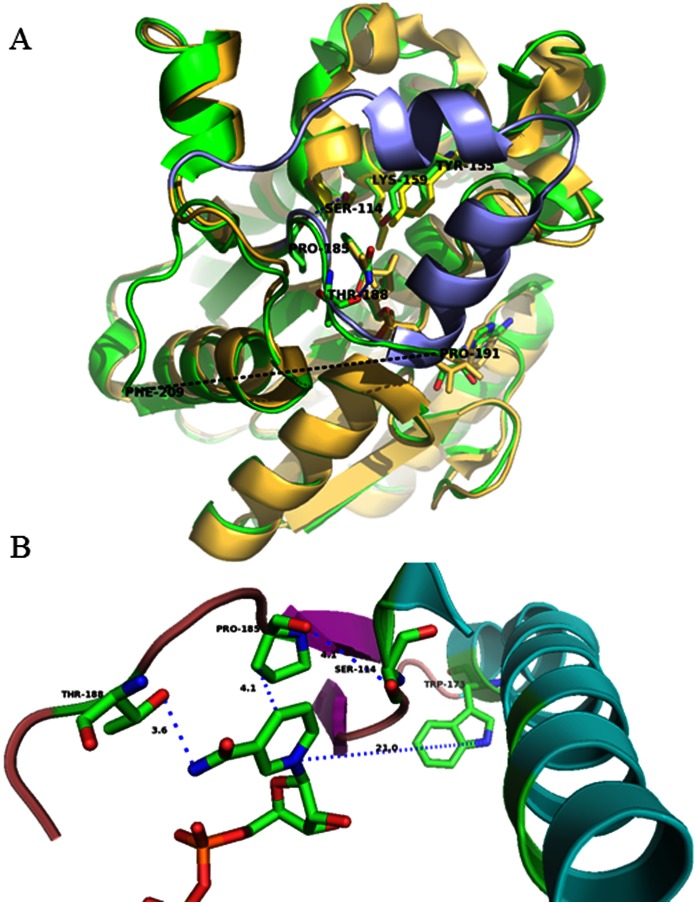
Figure 2. Structural alignment of P.s. 3α-hydroxysteroid dehydrogenase and C.t. 3α-HSD/CR.
A. Coenzyme NADH binding to P.s. 3α-HSD (yellow; pdb:2dkn) induces the loop-helix transition (blue) and an unresolved flexible loop (black dotted line) between P191 and F209 for the NAD+ bound C.t. 3α-HSD/CR (green; pdb:1fk8). NAD+ and key residues in the active site of 3α-HSD/CR are also labeled. (B) Close-up of the interaction of the substrate-binding loop with NAD+. The interactions of P185 at the hinge region with both S114 and nicotinamide ring, and T188 in the substrate-binding loop with the amide NH of NAD+ are shown as blue dotted lines. The distances from P185 to the nicotinamide ring of NAD+ and the hydroxyl group of T188 with the amide NH of NAD+ are 4.1 and 3.6 Å, respectively. The image was generated using the PyMOL program.