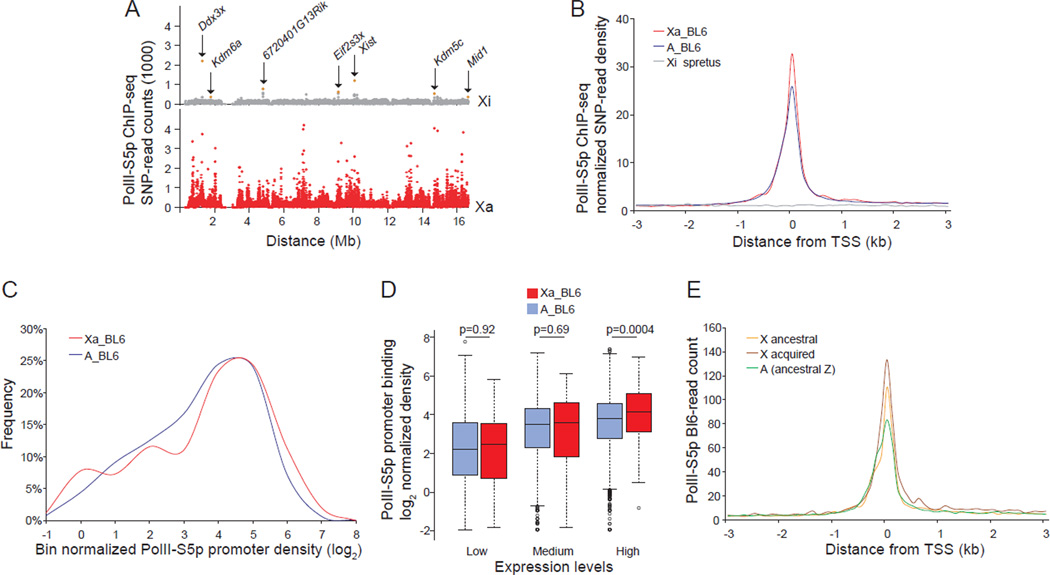
Figure 2. Higher PolII-S5p occupancy at expressed X-linked versus autosomal genes by allele-specific ChIP-seq analyses in brain.
(A–E) Allele-specific ChIP-seq in brain of an F1 mouse with the BL6 X chromosome active (Xa), and the spretus X inactive (Xi).
(A) Chromosome-wide allele-specific PolII-S5p occupancy profiles of the Xa and Xi are shown as SNP read counts in 10kb windows. Read counts for genes that escape X inactivation (Ddx3x, Kdm6a, 6720401G13Rik, Eif2s3x, Xist, Kdm5c, Mid1) are indicated (pink dots).
(B) Metagene analysis of PolII-S5p occupancy at the 5' end of expressed genes (≥1RPKM) on the Xa (Xa_BL6) (398 X-linked genes) and on the haploid set of autosomes (A_BL6) (11335 autosomal genes). SNP-read counts plotted 3kb up- and downstream of the TSS in 100bp windows.
(C) Percentage of expressed genes with PolII-S5p promoter occupancy is higher on the Xa (Xa_BL6) than on autosomal genes from the same haploid set (A_BL6) (p =0.007, KS (Kolmogorov–Smirnov)-test). Normalized promoter PolII-S5p density was calculated by averaging SNP-read counts in six 100bp windows around the TSS (peak region) for expressed genes, and normalizing by the median value of genomic DNA-SNP-read counts 3kb up- and downstream of the TSS.
(D) Box plots of normalized allele-specific promoter PolII-S5p density on expressed genes on the Xa compared to those on the haploid set of autosomes in each of the expression groups (low-, medium-, and high-expression). P values from Wilcoxon-test are shown.
(E) Metagene analysis of PolII-S5p allele-specific occupancy at the 5' end of expressed ancestral X-linked genes (282 genes) and acquired X-linked genes (155 genes), compared to ancestral autosomal genes (421 genes).
See also Figure S2.