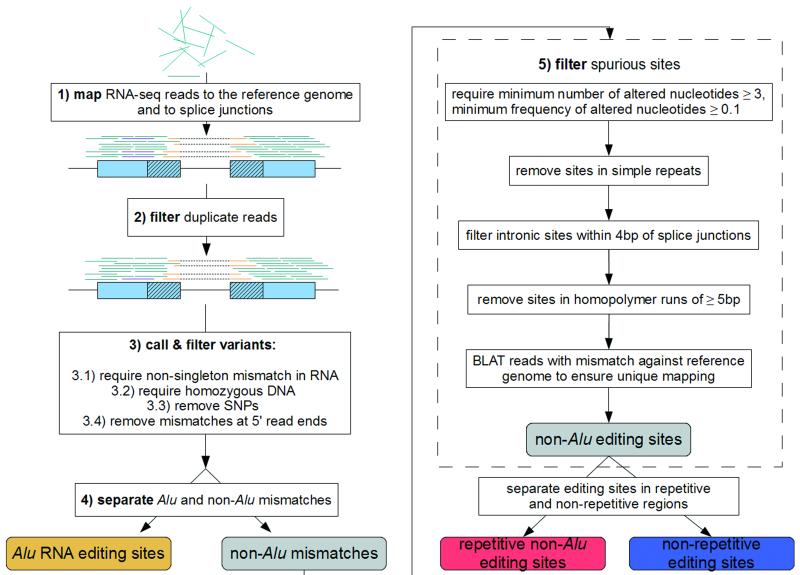
Figure 1. A computational framework to identify RNA editing sites in Alu and non-Alu regions.
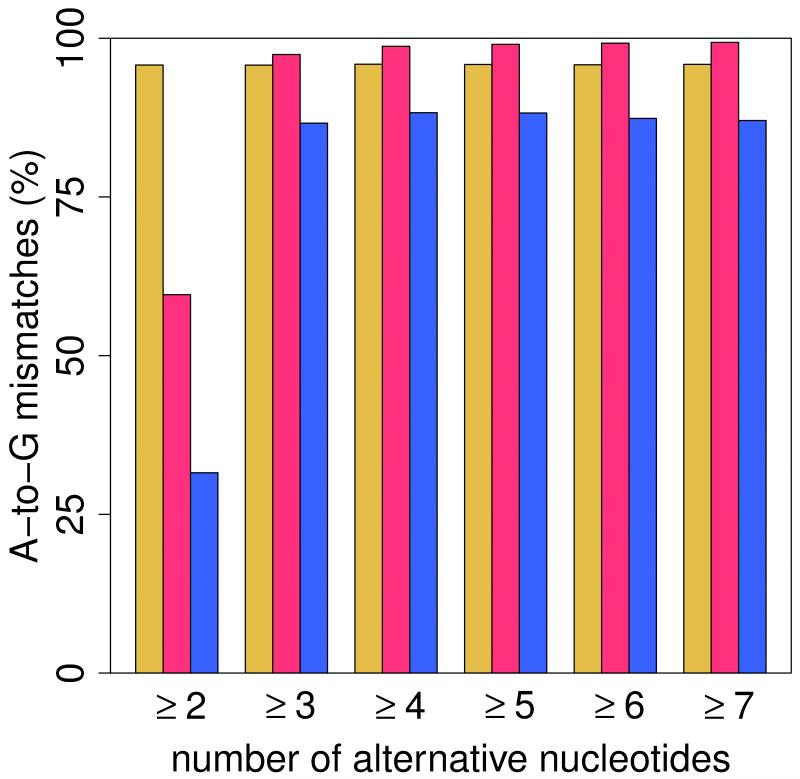
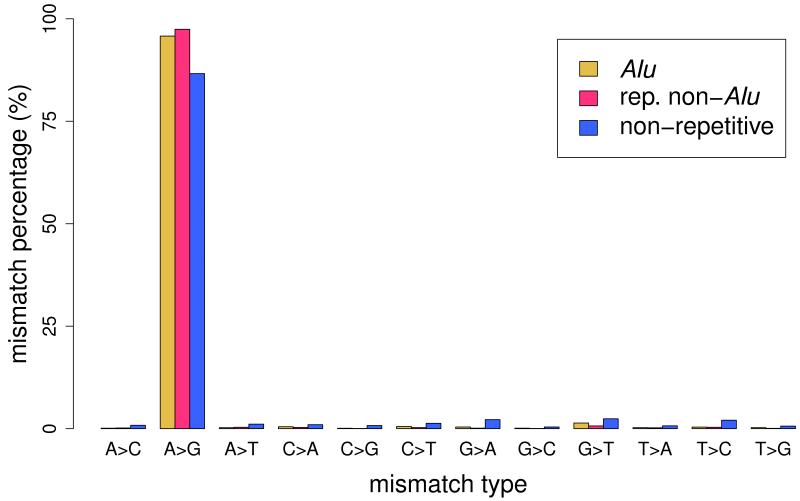
(a) Pipeline for the identification of RNA editing sites. RNA-seq reads (short lines) were mapped to the human reference genome (where blue reads map) and regions spanning all known splicing junctions (yellow lines separated by dashes). Boxes denote exons, and striped parts of two adjacent exons are joined together as the splicing junction sequence. (b) Relationship between the percentage of A-to-G mismatches and the minimum number of reads with altered nucleotides in Alu, repetitive non-Alu and nonrepetitive regions in GM12878. For all non-Alu sites, a minimum frequency of 10% for the RNA variant was required, whereas no minimum variant frequency was used for Alu positions. In non-Alu regions at least three variant nucleotides are required to achieve high specificity in RNA editing detection. (c) Percentage of all 12 mismatch types in GM12878.