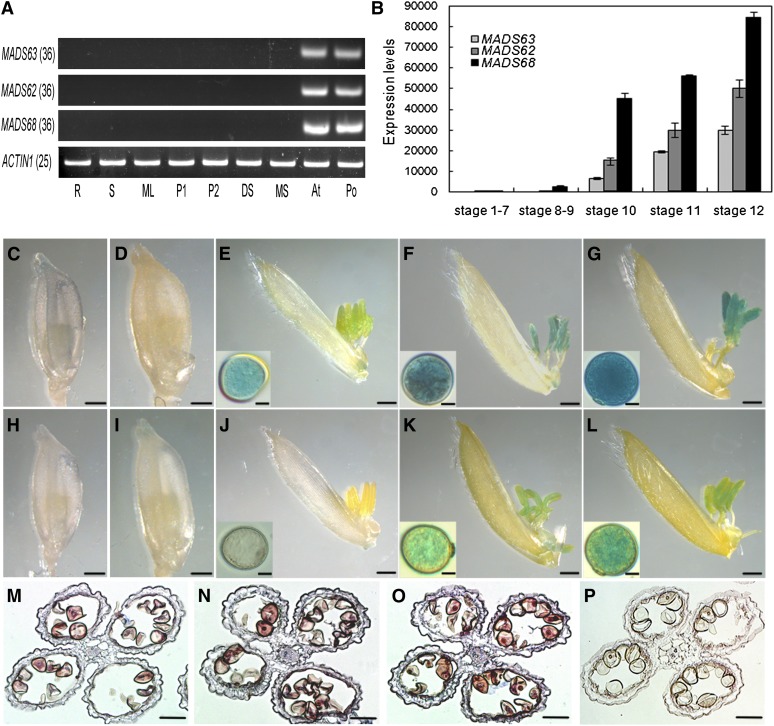
Figure 2.
Expression Pattern of Rice MIKC*-Type Genes.
(A) Expression analyses of three rice MIKC*-type genes in various tissues of rice by RT-PCR. Total RNA was isolated from of 7-d-old roots (R), 7-d-old stems (S), mature leaves (ML), 1- to 3-cm panicles (P1), 3- to 6-cm panicles (P2), a mixture of developing seeds (DS), a mixture of lemmas/paleas, lodicules, and pistils of mature spikelets (MS), anthers at stage 13 (An), and mature pollen (Po). ACTIN1 was used as control. The cycle number of PCR is shown in parentheses for each gene.
(B) Expression levels of three rice MIKC*-type genes in the anther at different developmental stages as revealed by qRT-PCR analysis. Total RNA was pooled from anthers of stage 1 to 7, stage 8 and 9, stage 10, stage 11, and stage 12. Data show the mean ± sd (n = 3).
(C) to (G) GUS staining analyses of florets and pollen grains at different developmental stages in MADS68pro:GUS transgenic plants. A stained floret with anther before stage 8 (C), at stage 9 (D), at stage 10 after the removal of lemma (E), at stage 11 after the removal of lemma (F), and at stage 12 after the removal of lemma (G). Insets ([E] to [G]) show a magnified pollen grain at the corresponding stage.
(H) to (L) GUS staining analyses of florets and pollen grains at different developmental stages in MADS63pro:GUS transgenic plants. A stained floret with anther before stage 8 (H), at stage 9 (I), at stage 10 after the removal of lemma (J), at stage 11 after the removal of lemma and insets (K), and at stage 12 after the removal of lemma (L). Insets ([J] to [L]) show a magnified pollen grain at the corresponding stage.
(M) to (P) In situ hybridization of MADS63 transcripts in anther at stage 11 (M), MADS62 transcripts in anther at stage 11 (N), MADS68 transcripts in anther at stage 11 (O), and negative control with sense probes of MADS68 (P).
Bars = 1 mm in (C) to (L), 10 µm in insets, and 50 µm in (M) to (P).