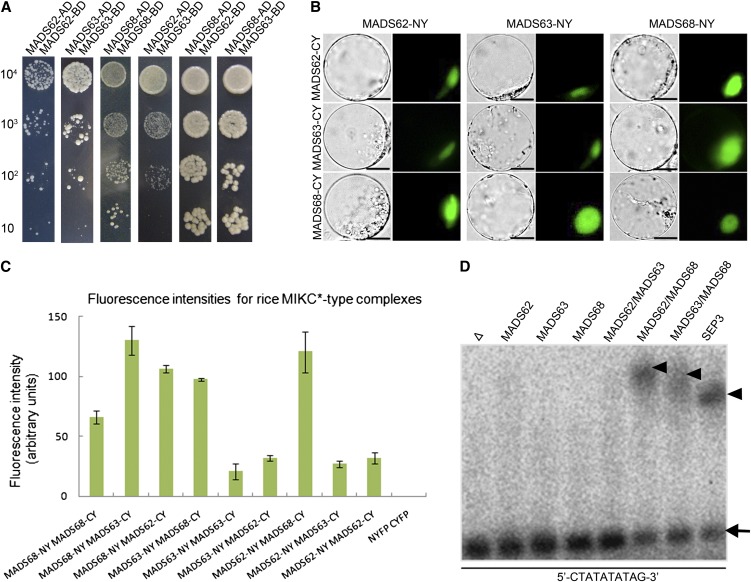
Figure 3.
Interaction Patterns and DNA Binding Ability of the Rice MIKC*-Type Proteins.
(A) Rice MIKC*-type protein interactions as revealed by Y2H. Each MIKC*-type protein was fused to the activation domain (AD) as a prey and the DNA binding domain (BD) as a bait. Serial dilutions of 104 to 101 AH109 cells containing different constructs combinations indicated were grown on the selective medium.
(B) BiFC yellow fluorescent protein fluorescence and bright-field images of rice protoplasts cotransfected with constructs encoding the indicated fusion proteins. Each MIKC*-type protein fused with the N-fragment or C-fragment of yellow fluorescent protein is labeled with its gene name followed by -CY and -NY, respectively. Bars = 10 µm
(C) Quantification of BiFC fluorescence intensities in transiently transfected rice protoplasts. Fluorescence intensity (arbitrary units) for each combination was determined to assess the strength of protein interactions. The mean and sd of six independent measurements are shown. Empty NYFP (N-terminal fragment of yellow fluorescent protein) and CYFP (C-terminal fragment of yellow fluorescent protein) were used as negative control.
(D) EMSA assay for rice MIKC*-type protein complexes binding N10-type CArG-box DNA. A probe containing an N10-type CArG-box (5′-CTATATATTAG-3′) was incubated with in vitro–translated MADS62, MADS63, MADS68, and combinations of these proteins. Free DNA is indicated by an arrow and shifted complexes by arrowheads. In vitro translation with SEPALLATA3 (SEP3) and an empty vector (Δ) served as positive and negative control, respectively.
[See online article for color version of this figure.]