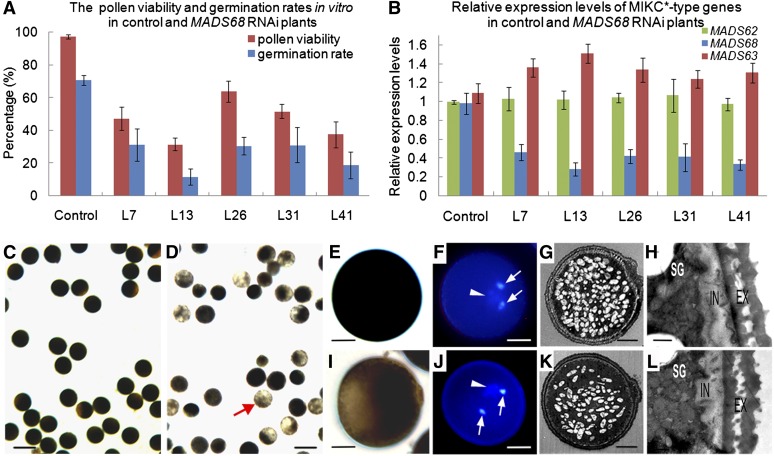
Figure 4.
Molecular Detection and Pollen Phenotypes of MADS68 RNAi Lines.
(A) The percentage of viable pollen grains and the germination rates in vitro of control plants and different MADS68 RNAi lines. The data show mean ± sd of three independent experiments.
(B) Relative expression levels of MADS68, MADS62, and MADS63 in control plants and five selected MADS68 RNAi lines (L7, L13, L26, L31, and L41). Total RNAs were pooled from anthers at stage 13. The samples were quantified by qRT-PCR using UBIQUITIN as a reference gene. The data are presented as mean ± sd (n = 3).
(C) to (L) Pollen phenotypes at the maturation stage. I2-KI staining showing pollen viability in control plant (C) and MADS68 RNAi L7 (D). The higher magnification of I2-KI staining pollen of control plant (E) and abnormal pollen of line MADS68 RNAi L7 (I). DAPI staining showing three nuclei of pollen grain in the control plant (F) and line MADS68 RNAi L7 (J). TEM showing starch accumulation in pollen of control plant (G) and abnormal pollen of line MADS68 RNAi L7 (K); wall structure in pollen of control plant (H) and abnormal pollen of line MADS68 RNAi L7 (L). Arrowheads indicate the vegetative nucleus, and arrows indicate sperm-cell nuclei in pollen of control plant (F) and abnormal pollen of line MADS68 RNAi L7 (J).
EX, extine; IN, intine; SG, starch granules. Bars = 100 µm in (C) and (D), 12.5 µm in (E), (F), (I), and (J), 5 µm in (G) and (K), and 0.5 µm in (H) and (L).