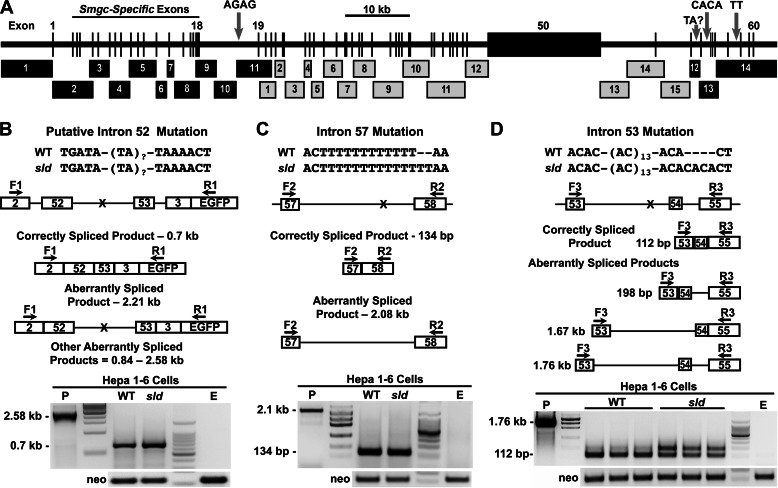
FIGURE 6.
Map of genomic sequencing strategy, resultant sequence variations and minigene splicing assays of the 3′-end mutations. A, genomic sequencing of Muc19/Smgc incorporated either cloned PCR products (black numbered boxes) or PCR products (gray numbered boxes). See “Experimental Procedures” for details. Three insertion mutations in introns 18, 53, and 57 are indicated by gray arrows. Because we were unable to sequence through TA repeats within intron 52, we label this region as a potential mutation site with unknown TA repeats. The central region of exon 50 is intractable to both cloning and sequencing because its central region contains ∼36 tandem repeats of 489 bp that encode Ser/Thr-rich sequences. B–D, minigene splicing assays of the 3′-end mutations. In each panel is shown the sequence variation between WT and sld mutant mice. Underneath is a map of the genomic sequence inserted into each minigene, along with positions of each sequence variation (X) and of forward (F) and reverse (R) primers used in subsequent PCRs of cDNA isolated from transfected Hepa 1–6 cells. Below each genomic map are analogous maps of correctly spliced products and of potential aberrantly spliced products. At the bottom of each panel are assay results, representative of at least two separate experiments with neo loading controls, an empty vector control (E) and a positive control (P) using DNA from the appropriate minigene. B and C, cells transfected with minigene DNA containing either intron 52 or 57 from each mouse genotype produced only a single PCR product representing the correctly spliced RNA. D, three preparations of cells transfected with 2 μg of minigene DNA from each genotype containing intron 53 displayed two products: the 112-bp correctly spliced product and a 198-bp aberrant product containing intron 54. The unmarked lanes show molecular size markers. The maps are not drawn to scale.