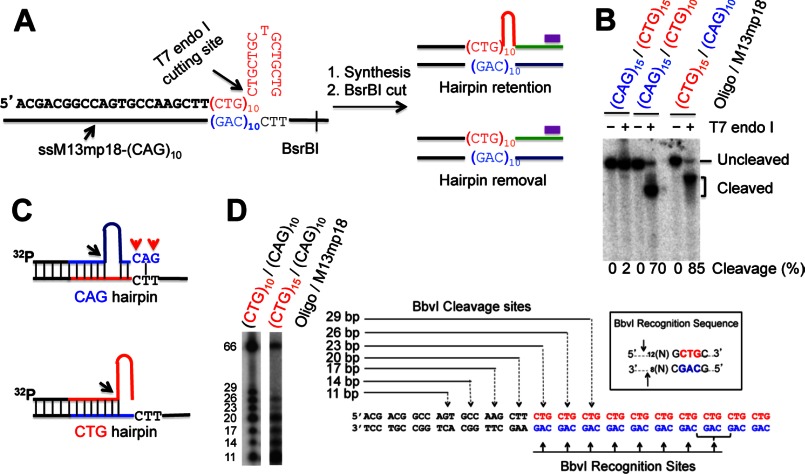
FIGURE 1.
DNA substrate and hairpin retention/removal assay. A, diagram of hairpin removal/retention assay by Southern blot analysis. The purple bar shows the 32P-labeled oligonucleotide probe, which specifically anneals to the newly synthesized strand near the BsrBI site. The complete primer sequence of a CTG hairpin substrate used in this study is also shown. B, substrate characterization. Each DNA substrate, which was 5′-32P-labeled in the primer strand, was digested with T7 endonuclease I, and the products were analyzed by denaturing polyacrylamide gel electrophoresis. The cleavage products were quantified. C, proposed structures for the CAG (upper) and CTG (lower) hairpin substrates. Black arrows point to the T7 cleavage sites, and red arrows indicate mismatches. The blue and red types/lines represent CAG and CTG repeats, respectively. D, digestion of a CTG hairpin by BbvI. The left panel shows the BbvI cleavage products of a (CTG)10/(CAG)10 homoduplex and a (CTG)15/(CAG)10 heteroduplex on a 15% denaturing polyacrylamide gel, and the right panel diagrams the recognition sites and the predicted cleavage sites of the (CTG)10/(CAG)10 homoduplex by BbvI.