Figure 4.
EBF1 Binds to the Gata3 Locus and Induces Repressive Histone Modifications
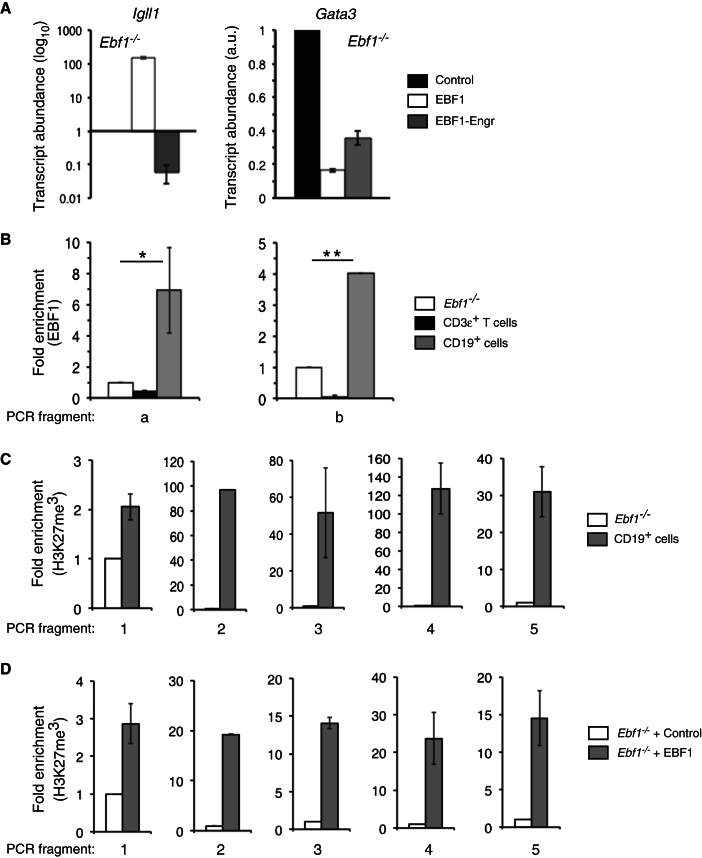
(A) Ebf1−/− lymphoid progenitors were transduced with a construct encoding an EBF1-Engrailed fusion protein, wild-type EBF1, or control virus. After 12 hr, GFP+ cells were harvested, cDNA prepared, and Igll1 or Gata3 mRNA levels determined with Taqman primer-probe sets. The data are normalized to transcript levels for either Igll1 or Gata3 in cells transduced with MigR1 control virus.
(B) ChIP via anti-EBF1 was performed with the indicated cell types including CD19+ BM lymphocytes and CD3ε+ thymocytes. qRT-PCR was performed to amplify immunoprecipitated DNA with flanking primers to sites a or b; detection signals were normalized to input DNA. Data are expressed as fold enrichment over Ebf1−/− cells, which served as negative control.
(C) H3K27me3 modifications within sites 1–5 in CD19+ BM lymphocytes relative to Ebf1−/− cells.
(D) Ebf1−/− cells were transduced with control MigR1 or MigR-EBF1 virus and plated on OP9 cells in presence of IL-7, FL, and SCF. 24 hr later, DAPI−GFP+ cells were sorted, fixed, and processed for ChIP. Relative H3K27me3 enrichment at sites 1–5 in EBF1 versus control-transduced samples is shown. Data are means ± SEMs of triplicate samples. Error bars indicate SEMs, *p < 0.01, **p < 0.001. Data are representative of four separate experiments.
See also Figures S3 and S4 and Table S1.