Figure 5.
Inhibition of EBF1 Binding to the Gata3 Locus Rescues T Cell Differentiation
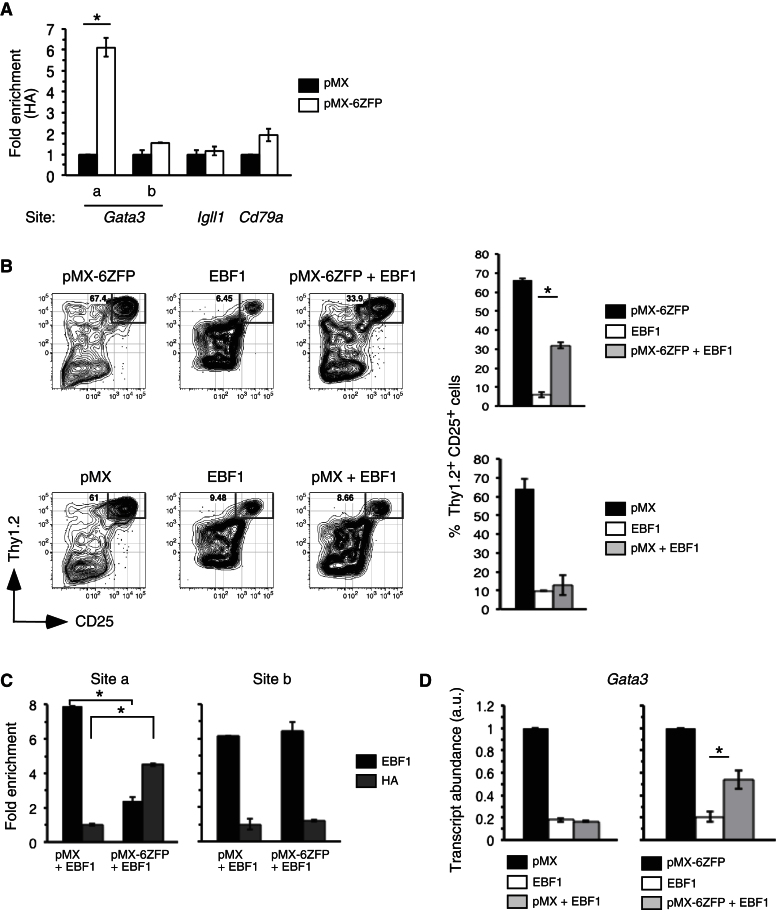
(A) ChIP experiments with HA antibodies were performed on GFP+ progenitors transduced 7 days previously with pMX-6ZFP or pMX (control) by site-specific PCR primers.
(B) Sorted e14.5 Pax5−/− fetal liver LSKs were cotransduced with pMX-ZF and MigY-EBF1 viruses and then added to OP9 stromal cells. pMX and MigY-EBF1 cotransduced cells served as control. After 24 hr, cells were washed and replated on OP9-DL4 stromal cells in fresh media supplemented with IL-7, FL, and SCF. On day 7, cultures were stained and analyzed for frequencies of Thy1.2+CD25+ cells among DAPI−CD45+ single- or double-transduced cells.
(C) Sorted e14.5 Ebf1−/− fetal liver LSKs were cotransduced with pMX and MigY-EBF1 or pMX-ZF and MigY-EBF1 viruses, added to OP9 stromal cells. After 7 days, viable YFP+GFP+ cells were sorted, fixed, and subjected to ChIP with either EBF1 or HA antibodies. Relative enrichment of EBF1 versus 6ZFP in transduced Ebf1−/− cells at “site a” is shown, with ChIP results for “site b” included as controls for EBF1 and 6ZFP occupancy. Data are expressed as fold enrichment over nontransduced Ebf1−/− cells.
(D) Single- or double-transduced cells from pMX and MigY-EBF1 transduced samples as well as pMX-ZF and MigY-EBF1 transduced samples were sorted for RNA 24 hr after infection and analyzed for relative expression of Gata3 by qRT-PCR. Data are means ± SEMs of triplicate samples. Error bars indicate SEMs, *p < 0.001. Data are representative of two separate experiments.