Abstract
A precise regulation of flowering time is critical for plant reproductive success, and therefore, a better understanding of the natural variation in genes regulating the initiation of the reproductive phase is required to develop well-adapted varieties. In both monocot and dicot species, the FLOWERING LOCUS T (FT) is a central integrator of seasonal signals perceived by the leaves. The encoded mobile protein (florigen) is transmitted to the apical meristem where it induces flowering. The FT homolog in barley (Hordeum vulgare L.), designated HvFT1, was shown to correspond to the vernalization locus VRN-H3, and natural alleles for spring and winter growth habit were identified. In this study, we demonstrate that the HvFT1 allele present in the barley genetic stock (BGS213) associated with a dominant spring growth habit carries at least four identical copies of HvFT1, whereas most barley varieties have a single copy. Increased copy number is associated with earlier transcriptional up-regulation of HvFT1 and a spring growth habit. This allele is epistatic to winter alleles for VRN-H1 and VRN-H2. Among accessions with one HvFT1 copy, haplotype differences in the HvFT1 promoter and first intron are also associated with differences in flowering time, which are modulated by genetic background. These different HvFT1 alleles can be used to develop barley varieties adapted to different or changing environments. Our results, together with studies of other wheat and barley flowering genes, show that copy number variation plays an important role in the regulation of developmental processes in the temperate cereals.
Keywords: Vernalization, Barley, Hordeumvulgare, Flowering, FT
Introduction
Plant reproductive success is highly dependent on a precise targeting of flowering time to a narrow seasonal window that maximizes resources for the developing seeds. In cereal crops, the correct targeting of this optimal reproductive period is translated into increased grain yields. A clear understanding of the natural variation in the genes that regulate flowering time is important to develop varieties adapted to different or changing environments.
In the temperate cereals, which include barley (Hordeum vulgare L.) and wheat (Triticum aestivum L.), the initiation of the reproductive phase is regulated by the integration of two main seasonal signals: photoperiod (day-length) and vernalization (extended exposures to low temperatures). Based on the response to photoperiod, barley varieties are divided into photoperiod sensitive (accelerated flowering under long days) and insensitive (limited response of flowering time to long days) classes. Based on the response to vernalization, barley varieties are divided into winter (vernalization accelerates flowering) and spring (early flowering irrespective of vernalization) classes.
Most of barley’s natural variation in photoperiod response is associated with allelic differences in the photoperiod genes PPD-H1 and PPD-H2. PPD-H1 encodes a pseudo-response regulator (PRR) protein that is part of the circadian clock (Turner et al. 2005) and promotes flowering under long days. Recessive mutations in the PPD-H1 gene reduce expression of HvFT1 and result in delayed flowering under long days (Hemming et al. 2008; Turner et al. 2005). The PPD-H1 gene acts in conjunction with CO-H1 (Campoli et al. 2012), which is one of the barley homologs of the Arabidopsis photoperiod gene CONSTANS (CO). In Arabidopsis, long days result in the stabilization of CO proteins, which up-regulate FT resulting in the acceleration of flowering (Corbesier and Coupland 2005). This function seems to be conserved in barley since overexpression of CO-H1 results in the up-regulation of HvFT1 and the acceleration of flowering (Campoli et al. 2012). The second photoperiod gene, PPD-H2 (HvFT3) is a paralog of HvFT1, but its effect on flowering is not as strong as HvFT1 (Kikuchi et al. 2009). The induction of flowering by PPD-H2 seems to be restricted to winter genotypes under non-inductive conditions (short days or long days without vernalization, Casao et al. 2011).
Natural variation in barley vernalization requirement is predominantly found in the vernalization loci VRN-H1, VRN-H2, and VRN-H3 (Dubcovsky et al. 2005; Fu et al. 2005; Takahashi and Yasuda 1971; Yan et al. 2005, 2006). The VRN-H1 gene is closely related to the Arabidopsis gene APETALA1, which encodes a MADS-box protein responsible for the transition of the shoot apical meristem (SAM) from the vegetative to the reproductive stage (Danyluk et al. 2003; Trevaskis et al. 2003; Yan et al. 2003). Deletions of regulatory regions in the VRN-H1 first intron result in a dominant spring growth habit (Fu et al. 2005; Hemming et al. 2009; von Zitzewitz et al. 2005). The second vernalization locus, VRN-H2, includes three closely related genes characterized by a putative zinc finger and a CCT-domain, designated as ZCCT genes (Yan et al. 2004). These genes function as flowering repressors and deletions of all three copies result in spring growth habit independently of the VRN-H1 alleles (Dubcovsky et al. 2005; Yan et al. 2004). Finally, VRN-H3 is a functional homolog of Arabidopsis flowering promoting gene FLOWERING LOCUS T (FT) (Yan et al. 2006) and will be referred hereafter as FT1 (HvFT1 in barley and TaFT1 in wheat).
In both monocot and dicot species, the signals from the vernalization and photoperiod pathways converge at the regulation of FT1, which is considered to be a central flowering integrator (Turck et al. 2008). When temperate cereals germinate during the fall, FT1 is repressed by VRN2, which competes with the photoperiod protein CO for the regulation of FT1 (Hemming et al. 2008; Trevaskis et al. 2007; Yan et al. 2006, Li et al. 2011). During the winter, vernalization up-regulates VRN1 (Oliver et al. 2009), which results in the repression of VRN2 in the leaves and the release of FT1 transcription in the spring (Loukoianov et al. 2005; Trevaskis et al. 2006; Hemming et al. 2008; Sasani et al. 2009; Chen and Dubcovsky 2012).
In Arabidopsis and rice, it has been demonstrated that FT encodes a mobile protein (florigen) that travels through the phloem and transmits signals perceived in the leaves to the SAM (Corbesier et al. 2007; Tamaki et al. 2007; Turck et al. 2008). Once FT arrives to the SAM it forms a complex with the bZIP transcription factor FD that physically interacts with the promoters of the meristem identity genes AP1 (Arabidopsis; Wigge et al. 2005 ), VRN1 (wheat; Li and Dubcovsky 2008) or FUL2 (rice; Tsuji et al. 2011), and up-regulates transcription to levels that induce the transition of the SAM to the reproductive stage. In rice, it has been shown that the FT-FD complex also includes 14-3-3 proteins (Taoka et al. 2011).
In barley and wheat, there are other FT-like genes (Faure et al. 2007; Kikuchi et al. 2009). However, FT1 shows the most robust induction of flowering when transformed into rice and wheat (Kikuchi et al. 2009; Yan et al. 2006) and is highly expressed under long days, which indicates that it is likely the key gene in the induction of flowering under long days.
In Arabidopsis, the FT promoter and first intron have been shown to contain cis-regulatory sites that are important for the transcriptional regulation of this gene (Adrian et al. 2010; Helliwell et al. 2006; Schwartz et al. 2009; Tiwari et al. 2010). However, the FT1 regulatory regions of barley and wheat are not as well characterized. Based on the few sequences available at the time of the cloning of HvFT1, Yan et al. (2006) found an association between growth habit and intron haplotypes. However, the sequencing of HvFT1 alleles from populations previously used to map QTL for flowering time, such as Dicktoo × Morex (Pan et al. 1994) and Sloop x Halcyon (Read et al. 2003; Hemming et al. 2008) failed to reveal any significant association between the intron one haplotypes and flowering time. Inconsistent results were also observed in recent surveys of HvFT1 allelic variation (Cuesta-Marcos et al. 2010; Casas et al. 2011). In this study, we demonstrate that these inconsistencies were generated by previously unknown copy number variation at the HvFT1 locus. We found that high HvFT1 copy number is associated with early flowering, and is epistatic to the vrn-H1 and Vrn-H2 alleles for winter growth habit (independently of PPD-H1). Finally, once we separated the effects of copy number variation, we were able to better characterize the effect of haplotype variation in HvFT1 regulatory regions on barley flowering time.
Materials and methods
Plant materials
Parental lines used in the different crosses were selected based on their HvFT1 alleles, including different combinations of promoter and first intron haplotypes. Their growth habits, vernalization and photoperiod alleles, and HvFT1 haplotypes are indicated in Table 1. All accessions except H. vulgare ssp. spontaneum Koch (OSU6, PBI004-7-0-015) belong to the sub-species H. vulgare ssp. vulgare. Hayakiso 2 and Iwate Mensury C (IMC) are commercial barley varieties from Japan and Igri is a variety from Germany, whereas E878 (Ethiopia) and U672 (Russia) are accessions received from the Okayama University barley collection (http://www.shigen.nig.ac.jp/barley/).
Table 1.
Parental lines used in the different segregating populations
| Line | Growth habit | PPD-H1 a | VRN-H1 b | VRN-H2 c | HvFT1 haploid copy number | HvFT1 haplo. abbr. | Promoter | Intron 1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 4 | 4 | 3 | 2 | 1 | 1 | 1 | 1 | 2 | 3 | |||||||
| 4 | 9 | 4 | 9 | 5 | 6 | 5 | 4 | 3 | 7 | 8 | |||||||
| 8 | 3 | 1 | 2 | 3 | 7 | 0 | 2 | 7 | 0 | 4 | |||||||
| BGS213 | S | ppd1 | vrn1 | Vrn2 | 4 | PE IAG | 4GC | G | A | C | T | i | T | C | C | A | G |
| IMC | S | Ppd1 | vrn1 | vrn2 | 1 | PE IAG | 4GC | G | A | C | T | i | T | C | C | A | G |
| Morex | S | ppd1 | Vrn1 | vrn2 | 1 | PE IAG | 4GC | G | A | C | T | i | T | C | C | A | G |
| E878 | S | ppd1 | Vrn1 | Vrn2 | 1 | PE ITC | 4GC | G | A | C | T | i | T | C | C | T | C |
| U672 | S | ppd1 | Vrn1 | Vrn2 | 1 | PE ITC | 4GC | G | A | C | T | i | T | C | C | T | C |
| Hayakiso2 | W | Ppd1 | vrn1 | Vrn2 | 1 | PE ITC | 4GC | G | A | C | T | i | T | C | C | T | C |
| Igri | W | Ppd1 | vrn1 | Vrn2 | 1 | PL ITC | 8GC | A | G | G | C | d | C | G | T | T | C |
| H. v. spont. | W | Ppd1 | vrn1 | Vrn2 | 1 | PL ITC | 8GC | A | G | G | C | d | C | G | T | T | C |
Growth habit, alleles at different vernalization genes, and HvFT1 copy number and haplotypes at the promoter (PE and PL) and intron one regions (IAG and ITC) are described for each line. Positions of the polymorphisms are reported as bp upstream of the start codon (promoter) or downstream of the start of intron one (intron) in Igri (i = insertion, d = deletion)
a PPD-H1: the recessive ppd1 allele is associated with photoperiod insensitivity and the dominant Ppd1 allele with photoperiod sensitivity
b VRN-H1: the recessive vrn1 allele is associated with vernalization requirement (winter growth habit) and the dominant Vrn1 allele is associated with the lack of vernalization requirement (spring growth habit)
c VRN-H2: the dominant Vrn2 allele is associated to vernalization requirement (winter growth habit) and the recessive vrn2 allele is associated with the lack of vernalization requirement (spring growth habit)
BGS213 is the designated genetic stock for the barley VRN-H3 (=Sgh3) allele associated with a dominant spring growth habit (Franckowiak and Konishi 1997). BGS213 is a line derived from the cross Tammi × Hayakiso 2 (GSHO 764) which has been selected for the dominant Vrn-H3 allele from Tammi (early flowering variety from Finland) and for recessive vrn-H1 and dominant Vrn-H2 alleles for winter growth habit (Takahashi and Yasuda 1971). Two additional sets of spring VRN-H3 isogenic lines were developed by backcrossing the VRN-H3 allele from Tammi into the winter varieties Hayakiso 2 and Dairokkaku 1 for eleven generations (Takahashi 1983), designated hereafter as Hayakiso 2-Tammi and Dairokkaku 1-Tammi.
Haplotypes
Based on a limited number of barley lines, it was initially hypothesized that haplotype variation in HvFT1 was associated to differences in flowering time (Yan et al. 2006). To facilitate the description of the different haplotypes used in this study, we assigned names to each of the haplotypes found in the promoter and intron one regions. The promoter haplotype present in E878, U672, BGS213, Hayakiso 2, and IMC is referred hereafter to as PE (promoter-early) whereas the promoter haplotype in Igri and H. vulgare ssp. spontaneum is referred to as PL (promoter-late). These two promoter haplotypes differ in nine linked SNPs and indels (Table 1). The two linked SNPs in the first intron are used to name the intron haplotypes as either ITC or IAG. Copy number variation is indicated separately in Table 1.
Segregating populations and growing conditions
A summary of the crosses and segregating populations included in this study is available in Table 2. The two crosses between BGS213 and winter lines H. vulgare ssp. spontaneum and Igri were published before (Yan et al. 2006) but are re-analyzed here in the light of the new copy number variation in HvFT1 presented in this study. Two additional crosses include the varieties Morex (Morex × H. vulgare ssp. spontaneum) and IMC (IMC × Hayakiso 2), which have identical HvFT1 haplotypes as BGS213 but different copy number. A third cross was made between BGS213 and IMC to test the effect of HvFT1 copy number variation on flowering time in lines with identical promoter and intron one haplotypes.
Table 2.
Segregating populations
| Parental lines | CNV | HvFT1 alleles | Number of plants tested (selected alleles) |
|---|---|---|---|
| BGS213 × H. vulgare ssp. spontaneum a | 4 × 1 | PE IAG × PL ITC | 72 (F2) |
| BGS213 × Igria | 4 × 1 | PE IAG × PL ITC | 96 (F2:3) |
| BGS213 × IMC | 4 × 1 | PE IAG × PE IAG | 164 (F2, Vrn-H2/−, Ppd-H1/−) |
| Morex × H. vulgare ssp. spontaneum | 1 × 1 | PE IAG × PL ITC | 81 (F2) |
| IMC × Hayakiso 2 | 1 × 1 | PE IAG × PE ITC | 70 (F2, Vrn-H2/−) |
| E878 × H. vulgare ssp. spontaneum | 1 × 1 | PE ITC × PL ITC |
47 (F3, Ppd-H1) 89 (F3, ppd-H1) |
| U672 × H. vulgare ssp. spontaneum | 1 × 1 | PE ITC × PL ITC | 42 (F2) |
| Hayakiso 2 × H. vulgare ssp. spontaneum | 1 × 1 | PE ITC × PL ITC | 125 (F2) + 134 (F2) |
Crosses made to study the effect of different combinations of HvFT1 haplotypes and copy number variation (CNV) on heading time
aYan et al. 2006
Finally, the effect of the different HvFT1 promoter haplotypes in varieties with identical HvFT1 copy number was tested in two F2 segregating spring x winter populations from crosses E878 × H. vulgare ssp. spontaneum and U672 × H. vulgare ssp. spontaneum and in one winter x winter population from the cross Hayakiso 2 × H. vulgare ssp. spontaneum. The effect of the different intron one haplotypes was only explored in the IMC × Hayakiso 2 segregating population.
All populations were produced and grown in greenhouse under long day (LD) photoperiod (15–16 h day length) generated by extending natural light conditions with supplementary lights as needed. Temperatures were held at non-vernalizing conditions (21–25 °C during the day and 12–18 °C during the night).
Markers
The Vrn-H1 and vrn-H1 alleles were identified using the UCW132 marker (Table 3), which detects a small indel near the end of the first intron that is linked to the larger functional deletion in the same intron. The two recessive vrn-H1 alleles in the Hayakiso 2 × H. vulgare ssp. spontaneum population were characterized using HvV1PromF2 and HvV1PromR2 primers that detect a promoter indel (Table 3). PPD-H1 alleles were identified using a Cleaved Amplified Polymorphic Sequence (CAPS) marker digested with restriction enzyme BstUI (Table 3; Turner et al. 2005). VRN-H2 was genotyped using primers VRN-H2aF and VRN-H2aR (Table 3), or when a codominant marker was necessary, with a marker for the tightly linked SNF2 gene (Table 3). The HvFT1 marker based on a 4-bp indel in the promoter was used for genotyping the populations segregating for the HvFT1 promoter haplotypes (Table 3; Yan et al. 2006), whereas the UCW133 marker was used to differentiate the intron haplotypes (Table 3).
Table 3.
Primer names, sequence, amplification temperatures, and amplification efficiencies
| Genotyping | |||||
|---|---|---|---|---|---|
| Gene | Primer name | Forward primer | Reverse primer | A.T. | Enzyme |
| Vrn-H1 | UCW132_F/R | TGTTTTGCAAACTATTTGACCAG | TAGCGCTCATACCGTTCAAG | 59° | – |
| vrn-H1 | HvV1PromF2/R2 | ACTTCACCCAACCACCTGAC | CTGGCGGTTGATCTTGTTCT | 55° | – |
| Vrn-H2 | VRN-H2a_F/R | CATGAAACAGCAGCTCCAGA | TTTGCCTCTCTCTCCTGCAT | 59° | – |
| SNF2 | SNF_F/R | TTGGTACTTGAATGCCTGAAAA | ATGGCACAACTTGGATTTGA | 60° | – |
| Ppd-H1 | PPD/H1/F/R | CTGAGCCTGAAGAGGTCGAG | GTGGCGGGAGGTTATCTCT | 57° | BstUI |
| HvFT1 Prom. | HvFT1F/R1 | ATGGACATGGAACCTGCCACT | TGGTGATGATGAGTGTTGCCC | 55° | – |
| HvFT1 Intron | UCW133_F/R | TGCACACACTTAGCGCAGTA | GCAGACCGTGGAACTCAACT | 55° | RsaI |
| EBmac603 | EBmac603F/R | ACCGAAACTAAATGAACTACTTCG | TGCAAACTGTGCTATTAAGGG | 56° | – |
| Bmag914 | Bmag914F/R | GGGCAATATACAGTTCAACTC | ATGAACTGGAGGCAGTAAATA | 57° | – |
| qRT-PCRa | ||||
|---|---|---|---|---|
| Gene | Primer name | Forward primer | Reverse primer | Efficiency % |
| UCW118 | CNV_UCW118_F1/R1 | CAGTAAGGCGAACCATGTCATC | TGCGCACCAACACAGAACA | 99.9 |
| FT1_Prom. | CNV_FT1p_F2/R2 | CGGCCGAGTCTGTGTGATCT | GGCATAAATCCCGCCTCTTT | 99.9 |
| FT1_ATG | CNV_FT1p_F5/R4 | TGTTCTAAGAAGGAAGGAGAAATGG | GAAGGTCACCCTGAGGTTGGT | 99.8 |
| FT1_Ex1 | CNV_FT1_F2/R2 | CGTACGTACACAATCACCACTATCTAATG | GAGAGCCCGATCGTGCAT | 99.8 |
| FT1_Ex3 | CNV_FT1_F4/R3 | GCAGGTTGGTGACAGATATCCC | GGAAGAGCACGAGCACGAA | 89.3 |
| UCW123 | CNV_UCW123_F1/R1 | ACTGCAAGAGCTACAGCCTTCA | GTCACCGGCAGCAAGATCTAG | 99.9 |
| UCW120 | CNV_UCW120_F1/R1 | GCGACGACCAGTAAAAAATGC | CCGTTTCCGTGGATGGAA | 99.9 |
| SNF2 | CNV_SNF2_F1/R1 | ATTACCGCTCTGCTGTCGCGATTA | AAATGTGGCTCTGAAGGTGTTGGC | 93.4 |
aPCR program:(95° 20 s) × one cycle, (95° 3 s, 60° 30 s) × 40 cycles
Determination of copy number variation (CNV) by quantitative PCR
Genomic DNA was extracted using the CTAB (cetyltrimethylammonium bromide) extraction method (Murray and Thompson 1980). Samples were treated with RNAse A, and a phenol:chloroform:isoamyl alcohol purification step was used to remove possible RNA contamination, which might interfere with normalization. DNA concentration was normalized using a Nanodrop instrument (Thermo Fisher Scientific, Waltham, MA) to a concentration of 20 ng/μl, and 1 μl was used for each 20 μl Fast Sybr®Green reaction. These reactions were performed on an AB7500 Fast Real-Time PCR System (Applied Biosystems by Life Technologies, Grand Island, New York), using identical programs for all primer pairs ((95° 20 s) × one cycle, (95° 3 s, 60° 30 s) × 40 cycles). The parental lines were tested with various pairs of primers in the HvFT1 region (Table 3), and a pair of primers in the SNF2 control gene, which has been shown before to have a single copy in the barley genome (Yan et al. 2002). The 2−ΔΔCт method was used to estimate copy number (Weaver et al. 2010), with Morex as a calibrator and CT = threshold cycle. We first calculated ΔMorex = Morex–HvFT1 CT−Morex–SNF2 CT and ΔTarget = Target–HvFT1 CT−Target–SNF2 CT, and then calculated the difference between the two as −ΔΔCT = ΔMorex−ΔTarget. Previous sequencing of Bacterial Artificial Chromosomes (BACs) containing HvFT1 has shown that only one copy of this gene is present in Morex, making it a good calibrator variety (Yan et al. 2006). Table 3 describes the primers used for quantitative PCR and their efficiency.
The first HvFT1 primer pair (CNV_FT1_F2 and CNV_FT1_R2) was designed on the border between the first exon and the first intron. This region was selected because it includes several SNPs that differentiate HvFT1 from other members of the FT family. Copy number in the recombinant BGS213 × IMC F2 lines was determined using these primers.
To determine the borders of the duplication in the BGS213 lines, additional primers were designed inside and flanking the HvFT1 gene. In the HvFT1 promoter region, a set of primers was designed 656–727 bp upstream of the start codon (CNV_FT1p_F2 and CNV_FT1p_R2). Another set of primers amplified a region from a 22-bp upstream to 97-bp downstream of the start codon (CNV_FT1p_F5 and CNV_FT1p_R4). Within the gene, an additional set of primers was designed at exon three to determine if the complete gene was duplicated (CNV_FT1_F4 and CNV_FT1_R3, 778-902 bp downstream from the start codon). Markers were also designed outside the HvFT1 gene. One pair of primers was designed for gene UCW123, located 6.6 kb downstream from HvFT1 on Morex BAC clone 440G4 (GenBank DQ900686, CNV_UCW123_F1 and CNV_UCW123_R1). The nearest known genes on flanking BACs 455J22 (GenBank DQ900687) and 761F04 (GenBank DQ900685, Yan et al. 2006) were also tested, including UCW120 (CNV_UCW120_F1 and CNV_UCW120_R1) and UCW118 (CNV_UCW118_F1 and CNV_UCW118_R1).
HvFT1 expression profiles
To test the effect of the dosage of the duplicated HvFT1 locus, the isogenic lines of Hayakiso 2 with and without the Tammi HvFT1 allele were intercrossed and the F2 lines were genotyped for HvFT1 and then tested for HvFT1 expression by quantitative RT-PCR (qRT-PCR) with Fast Sybr®Green Master Mix on the AB7500 Fast Real-Time PCR System (Yan et al. 2006). ACTIN was used as an endogenous expression control (Trevaskis et al. 2006).
Results
Variation in flowering time among lines with identical HvFT1 haplotypes
The HvFT1 sequences from Morex, IMC, and BGS213 are identical (Table 1). However, segregating populations including the first two varieties (Morex × H. vulgare ssp. spontaneum and IMC × Hayakiso 2) showed that plants with winter alleles for the VRN-H1 and VRN-H2 genes flowered very late independently of HvFT1. This observation was inconsistent with the results from Yan et al. (2006), which have previously shown that the HvFT1 allele from BGS213 was sufficient to confer a spring growth habit when introgressed into winter barley varieties. To test if additional polymorphism were present among these three HvFT1 alleles, we sequenced a 9,250-bp region starting 1,499-bp upstream of the start codon and ending 6,690-bp downstream of the stop codon. No polymorphisms were detected in the HvFT1 coding and flanking sequences of Morex, IMC, and BGS213. The segregating populations including these varieties are described in detail below.
Morex (PE IAG) × H. vulgare ssp. spontaneum (PL ITC)
This population of 81 F2 plants segregated for the VRN-H1, VRN-H2, and HvFT1 loci. To study the effect of the HvFT1 alleles in a winter background, we first selected plants homozygous for vrn-H1 and homozygous or heterozygous for the Vrn-H2 alleles for winter growth habit. The 11 selected plants carrying these alleles flowered relatively late (91–106 days after sowing) and showed no significant differences between the different HvFT1 alleles (P = 0.32), even when the 11 plants were divided in photoperiod sensitive and photoperiod insensitive groups.
IMC (PE IAG) × Hayakiso 2 (PE ITC)
A similar result was observed in a population of 92 F2 lines from the cross between the photoperiod sensitive varieties IMC and Hayakiso 2. IMC is a spring barley variety that carries an HvFT1 allele with identical sequence to the BGS213 allele and a vrn-H2 allele for spring growth habit, whereas Hayakiso 2 is a winter variety (Table 1). In the F2 segregating population, 22 plants homozygous for the IMC vrn2 allele were identified using the tightly linked SNF2 molecular marker and were discarded. The remaining 70 plants, carrying vrn-H1 and Vrn-H2 alleles for winter growth habit, flowered relatively late (100–250 days, average 167 days) and showed no significant differences between the different HvFT1 alleles (P > 0.05). Plants homozygous for the functional Vrn-H2 allele flowered 29 days later than the plants carrying the heterozygous allele (P = 0.02) suggesting some dosage effect of VRN-H2. No significant interaction was detected between HvFT1 and VRN-H2 (P = 0.74).
The results from the IMC × Hayakiso 2 and Morex × H. vulgare ssp. spontaneum populations suggest that, in spite of their identical sequence, the HvFT1 alleles present in Morex and IMC have a smaller effect on inducing flowering than the HvFT1 allele present in BGS213. To test this hypothesis, we developed a third population segregating for the HvFT1 alleles of BGS213 and IMC, which have identical sequence and contrasting phenotypes.
BGS213 (PE IAG) × IMC (PE IAG)
Since no polymorphisms were found between the two parental lines in HvFT1 or in the genes tightly linked to HvFT1 (Yan et al. 2006), we screened linked SSR markers and found Bmag914 and EBmac0603 (GrainGenes database, http://wheat.pw.usda.gov/) to be polymorphic between BGS213 and IMC. These two markers flanking the HvFT1 locus were located approximately 14 cM apart in this mapping population.
The complete segregating population was genotyped with the HvFT1 flanking SSR markers and with markers for VRN-H2 and PPD-H1 genes. To focus on the effect of HvFT1, plants homozygous for the vrn-H2 allele for spring growth habit or for the photoperiod insensitive allele (ppd-H1) were eliminated. The remaining 167 F2 lines showed a highly significant (P < 0.0001) effect on flowering associated with the SSR markers flanking the HvFT1 locus. The HvFT1 locus alone explained 96 % of the variation in flowering time among the selected 167 plants, indicating that no other major flowering gene was segregating in this selected sub-population.
Plants heterozygous or homozygous for the BGS213 HvFT1 allele showed a spring growth habit (33 days to 76 days from sowing to heading time) whereas plants homozygous for the IMC SSR markers flanking the HvFT1 locus showed no signs of flowering 125 days after planting, when the experiment was terminated. This population showed a clear 3:1 ratio between spring and winter plants (122 spring/45 winter, χ 2 P = 0.56). In summary, these results confirmed that the largest differences in flowering time in this segregating population were linked to the HvFT1 region in spite of the identical HvFT1 sequences between BGS213 and IMC.
Variation in HvFT1 copy number
Possible explanations for the previously described differences in flowering time include an unknown gene tightly linked to HvFT1, a regulatory sequence beyond the HvFT1 sequenced region or copy number variation (CNV) in HvFT1. To test the last hypothesis, we determined the number of HvFT1 copies in BGS213, IMC, Morex, and other accessions used as parental lines in different segregating populations.
DNAs were extracted from approximately 10 plants from each variety, and HvFT1 copy number was determined by quantitative PCR for the exon one region (Table 3) using the single copy gene SNF2 as an internal reference and Morex as a calibrator (see Materials and Methods). The results indicated that BGS213 has four to five copies of HvFT1 whereas IMC has a single copy (Fig. 1). The increase of HvFT1 copy number found in the BGS213 allele was confirmed in the two backcross substitution lines of the Tammi HvFT1 allele into the winter varieties Hayakiso 2 and Dairokkaku 1, which also showed four to five copies of HvFT1 each. All other varieties tested in this experiment (E878, H. vulgare ssp. spontaneum, Hayakiso 2, Golden Promise, Igri, U672 and Morex) showed only one copy of HvFT1.
Fig. 1.
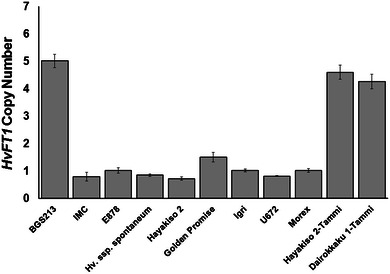
HvFT1 haploid copy number is increased in BGS213. Copy number was determined using the 2−ΔΔCT method (Weaver et al. 2010) and primers for the first exon (FT1_Ex1, Table 3). The single copy gene SNF2 was used as internal control and the variety Morex as calibrator (see “Materials and methods”). Averages and standard errors of the means are based on 7–10 biological independent DNA extractions
To test if the multiple HvFT1 copies were linked, we estimated HvFT1 copy number with the same exon one primers in 45 BGS213 x IMC F2 lines selected for recombination between the HvFT1 flanking markers Bmag914 and EBmac0603. The lines that flowered earlier (30–39 days) showed the highest average copy number (~5 copies), whereas the lines that flowered later (>110 days) showed an average of one HvFT1 copy. Plants with intermediate flowering times are likely heterozygous since they showed an intermediate copy number (~3 copies). These results confirmed that HvFT1 copy number co-segregates with flowering time (Fig. 2), and that the different HvFT1 copies are linked.
Fig. 2.
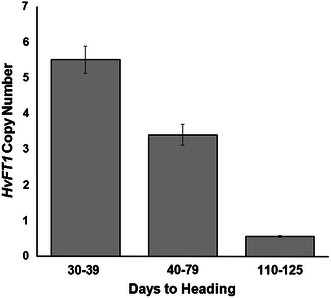
Co-segregation of HvFT1 copy number and heading time in 45 F2 lines from the BGS213 × IMC population selected for recombination between SSR markers flanking the HvFT1 gene. Late flowering lines showed a single HvFT1 copy, lines with intermediate flowering showed on average ~3 copies, and early flowering lines showed an average of ~5 copies. Haploid copy number was determined as described in Fig. 1
We performed an additional experiment to determine the extension of the duplicated region. Primer pairs within loci UCW118 and UCW120 located in Morex’s HvFT1 flanking BACs 455J22 and 761F04 (Yan et al. 2006), showed no evidence of duplication in BGS213 (Fig. 3). These results indicated that the duplication of the HvFT1 region did not include the adjacent BACs. Therefore, we designed additional primers within the Morex BAC 440G04, which includes the HvFT1 gene. The primer set for the HvFT1 promoter region located 655–700 bp upstream of the ATG start site showed no increase in copy number (Fig. 3). Since the start codon region shows four to five copies, the duplicated region must start within the 600-bp region of the HvFT1 promoter upstream of the start codon. The primers for the third exon of HvFT1 and for the UCW123 marker located 6.6-kb downstream from HvFT1 showed that the duplicated region extended beyond the coding region of HvFT1 and into this marker (Fig. 3).
Fig. 3.
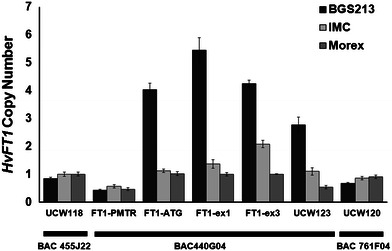
Extension of the HvFT1 duplicated region in BGS213. Primers in the flanking BACs (Yan et al. 2006) and the HvFT1 promoter (FT1-PMTR, 655–700 bp upstream of start codon) showed no evidence of duplication. Primers flanking the start codon (FT1-ATG), in the first exon (FT1-ex1), in the third exon (FT1-ex3) and in the UCW123 marker 6.6-kb downstream of the stop codon showed evidence of duplication. Averages and standard errors of the means are based on 7–10 independent DNA extractions
Differences in HvFT1 expression in alleles with different copy number
To study the relationship between HvFT1 copy number variation and expression, we crossed Hayakiso 2 with its near isogenic line containing the Tammi allele (Hayakiso 2-Tammi), selected eight plants homozygous for each of the parental alleles and eight heterozygous plants from the segregating F2 population, and characterized HvFT1 expression profiles in the leaves by qRT-PCR.
Hayakiso 2-Tammi lines showed earlier expression of HvFT1, with significantly higher transcript levels of HvFT1 expression than the two other genotypes just 2 weeks after planting (three leaf stage; Fig. 4, P = 0.002). Plants homozygous for the Tammi HvFT1 allele flowered around seven to 8 weeks, after which HvFT1 transcripts fell back to lower levels. The heterozygous plants showed an increase of HvFT1 transcript levels at the last time point of the experiment (11 weeks), when approximately one-third of the plants had already headed. At this same time point, plants homozygous for the HvFT1 alleles from Hayakiso 2 showed no detectable levels of HvFT1 and no signs of flowering induction (Fig. 4). These results show that the transcript levels of HvFT1 are affected by copy number, with larger copy number resulting in earlier HvFT1 expression and early flowering.
Fig. 4.
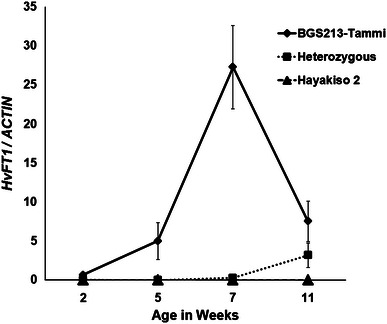
HvFT1 expression profiles correlate with copy number in a segregating population from the cross between isogenic lines Hayakiso 2 and Hayakiso 2-Tammi. Plants homozygous for the Tammi allele (black solid line with black diamonds) showed earlier induction of HvFT1 than those homozygous for Hayakiso 2 allele (dashed black line with triangles). In the heterozygotes plants (dotted black line with squares) HvFT1 transcript levels started to be induced by the end of this time course experiment. Bars are plus-minus one standard error of the means
Association between differences in HvFT1 haplotypes and heading time
Since HvFT1 copy number differences have such a large effect on flowering time, the effect of the different HvFT1 haplotypes needs to be tested using varieties with the same copy number.
Only one population was available to study the effect of the HvFT1 first intron haplotypes in lines with a single HvFT1 copy and identical HvFT1 promoter haplotypes (Table 1).
IMC (PE IAG) × Hayakiso 2 (PE ITC)
Both IMC and Hayakiso 2 are photoperiod sensitive, and carry a single copy of HvFT1 with the same promoter haplotype, but they differ in their first intron haplotypes (Table 1). Plants homozygous for the IAG allele flowered on average 10 and 13 days later than the plants heterozygous and homozygous for the ITC allele, respectively. However, these differences were not significant (P = 0.19 within the VRN2 heterozygous class) likely due to the large variability of late flowering plants (100–250 days) that resulted in a reduced statistical power. The late flowering observed in all plants indicates that none of the HvFT1 alleles segregating in this population was epistatic to the VRN-H1 and VRN-H2 alleles for winter growth habit.
Three additional populations were used to study the effect of the HvFT1 promoter haplotypes in lines with a single HvFT1 copy (Fig. 3) and identical HvFT1 intron one haplotypes (Table 1).
E878 (PE ITC) × H. vulgare ssp. spontaneum (PL ITC)
This F2 population segregated for HvFT1, VRN-H1 and PPD-H1. To simplify the analysis, two separate F3 segregating populations were derived in which the photoperiod sensitive (Ppd-H1) and photoperiod insensitive (ppd-H1) alleles were fixed. The ANOVA models including the VRN1 and HvFT1 loci, and their interactions explained 92 and 94 % of the variation in heading time in the photoperiod sensitive and insensitive subpopulations, respectively, which indicates that these two genes account for most of the variation in heading time in these two sub-populations. Both sub-populations showed highly significant interactions between the VRN-H1 and HvFT1 loci (P < 0.0001) and therefore, the effects of the HvFT1 alleles on heading time are described separately for the recessive and dominant (homozygous plus heterozygous) VRN-H1 classes.
The statistical analysis of the photoperiod sensitive F3 family (47 plants, Fig. 5a) showed significant effects of the HvFT1 alleles within both the dominant Vrn-H1 class (9.5 days, P = 0.0031) and the recessive vrn-H1 class (39 days, P < 0.0001). Here, the differences in heading time were approximately fourfold larger within the recessive vrn-H1 class than within the dominant Vrn-H1 class. The HvFT1 allele from E878 was associated in both cases with early flowering.
Fig. 5.
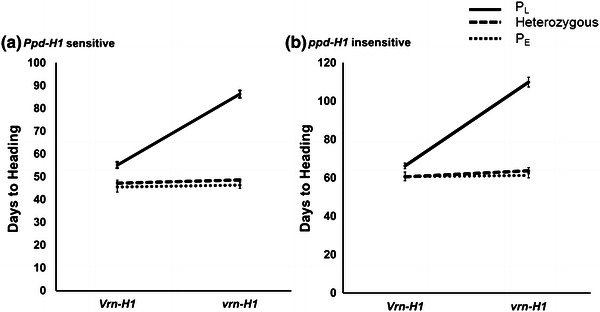
VRN-H1 and HvFT1 alleles show significant interaction in E878 (PE ITC) x H. vulgare ssp. spontaneum (PL ITC) populations segregating for the HvFT1 promoter haplotypes. a Photoperiod sensitive F3 family, b photoperiod insensitive F3 family. The lack of parallelism between lines reflects the significant interaction between VRN-H1 and HvFT1 alleles (larger effect of the HvFT1 alleles within the vrn-H1 class than within the Vrn-H1 class). Bars are plus-minus one standard error of the means
The plants from the photoperiod insensitive F3 family (89 plants, Fig. 5b) flowered on average 20 days later than the photoperiod sensitive F3 family (Fig. 5a). As in the photoperiod sensitive plants, the effect of the HvFT1 promoter haplotypes was larger among the plants homozygous for the recessive vrn-H1 allele (48.5 days, P < 0.0001) than among those homozygous or heterozygous for the dominant Vrn-H1 allele (5.5 days, P = 0.0002). As in the previous family, the HvFT1 PE haplotype from E878 was associated with early flowering.
The reciprocal epistatic effect was observed when the effect of the VRN-H1 alleles was studied within each of the HvFT1 classes. No significant differences in heading time were detected between VRN-H1 alleles among the plants homozygous for the HvFT1 PE promoter allele, but large differences were observed among the plants homozygous for the HvFT1 PL promoter allele, both in the photoperiod sensitive (30 days, P < 0.0001) and the photoperiod insensitive sub-populations (43.6 days, P < 0.0001).
U672 (PE ITC) × H. vulgare ssp. spontaneum (PL ITC)
Similar results were observed in an additional small F2 population (42 plants) segregating for the promoter haplotypes. In this population, the effect of the HvFT1 alleles was also larger within the plants homozygous for the vrn-H1 allele for winter growth habit (17 days difference, P = 0.008) than among the plants including a dominant Vrn-H1 allele in homozygous or heterozygous state (6 days difference, P = 0.10). In both classes, the PE allele was associated with early flowering.
In both E878 and U672 populations, the HvFT1 PE ITC haplotype was associated with early flowering and was epistatic to the winter VRN-H1 and VRN-H2 alleles. However, the same PE ITC haplotype is also present in the variety Hayakiso 2 (Table 1), which has a winter growth habit. To test if this effect was caused by unlinked epistatic genes in Hayakiso 2 or by a specific characteristic of the HvFT1 region in Hayakiso 2, we crossed this variety with H. vulgare ssp. spontaneum to generate a third population segregating for the HvFT1 promoter haplotype. These two varieties are photoperiod sensitive and have a winter growth habit.
Hayakiso 2 (PE ITC) × H. vulgare ssp. spontaneum (PL ITC)
This population was characterized in two separate experiments performed in California (125 F2) and Oklahoma (134 F2). The effect of the HvFT1 alleles was consistent across experiments (no significant experiment × HvFT1 interaction), so the results from the two experiments were analyzed in a single factorial ANOVA including experiment, VRN-H1 and HvFT1 as factors. This analysis showed significant differences in heading time between the two recessive vrn-H1 alleles (P < 0.0001) and also between the HvFT1 PE and PL alleles (P < 0.0001), but no significant interaction between the two loci (P = 0.36). On average, F2 plants carrying the vrn-H1 allele from H. vulgare ssp. spontaneum (average 106 days after sowing) flowered 31 days earlier than those carrying the corresponding allele from Hayakiso 2 (135 days after sowing, Table 4). These results indicated that even though both varieties have recessive vrn-H1 alleles for winter growth habit, the vrn-H1 region from Hayakiso 2 is associated with a stronger vernalization requirement than the one from H. vulgare ssp. spontaneum. The earlier vrn-H1 allele was dominant, since the difference in heading time between plants heterozygous and homozygous for the H. vulgare ssp. spontaneum VRN1 allele was less than 1 day (Table 4). Surprisingly, plants carrying the HvFT1 allele from Hayakiso 2 (PE promoter) were on average 10 days later than the heterozygous plants and 14 days later than the plants homozygous for the HvFT1 allele from H. vulgare ssp. spontaneum (PL promoter), and the differences were highly significant (P < 0.0001, Table 4).
Table 4.
Hayakiso 2 (PE ITC) × H. vulgare ssp. spontaneum (PL ITC): average heading time plus-minus standard error of the means
| VRN-H1 allelea | HvFT1 allele | Heading time (days) | ±SEM | Tukey |
|---|---|---|---|---|
| H. v. spontaneum | H. v. spontaneum | 97.9 | 4.1 | a |
| H. v. spontaneum | Heterozygous | 103.5 | 3.7 | ab |
| H. v. spontaneum | Hayakiso 2 | 115.3 | 4.3 | b |
| Avg. H. v. spontaneum vrn-H1 | 105.6 | a | ||
| Heterozygous | H. v. spontaneum | 98.1 | 3.1 | a |
| Heterozygous | Heterozygous | 106.9 | 2.5 | ab |
| Heterozygous | Hayakiso 2 | 109.5 | 3.1 | b |
| Avg. heterozygous vrn-H1 | 104.8 | a | ||
| Hayakiso 2 | H. v. spontaneum | 124.7 | 3.7 | a |
| Hayakiso 2 | Heterozygous | 140.9 | 3.8 | b |
| Hayakiso 2 | Hayakiso 2 | 137.9 | 3.4 | b |
| Avg. Hayakiso 2 vrn-H1 | 134.5 | b | ||
Different letters in the last column indicate significant differences between the mean values using the Tukey test (P < 0.05)
aBoth VRN-H1 alleles in this population are recessive vrn-H1
Discussion
HvFT1 increases in copy number are associated with a spring growth habit
Previous studies demonstrated that the BGS213 HvFT1 allele is sufficient to confer spring growth habit and therefore that it is epistatic to the VRN-H1 and VRN-H2 alleles for winter growth habit (Takahashi and Yasuda 1971; Yan et al. 2006). In the two F2 populations reported by Yan et al. (2006) (BGS213 × H. vulgare ssp. spontaneum and BGS213 × Igri), plants heterozygous or homozygous for the BGS213 HvFT1 allele had a spring growth habit (average heading time 40 and 59 days, respectively) and the plants homozygous for the H. vulgare ssp. spontaneum or Igri alleles showed very late flowering or failed to flower (average heading time 106 and >130 days, respectively; Yan et al. 2006). In addition, the introgression of the HvFT1 early flowering allele from Tammi into the winter-photoperiod sensitive varieties Hayakiso 2 and Dairokkaku 1 also resulted in spring growth habit (Yan et al. 2006). Results from the BGS213 × IMC population in this study confirmed the previous results. Taken together, these experiments indicated that the epistatic effect of the BGS213 HvFT1 allele on the VRN-H1 and VRN-H2 alleles for winter growth habit is effective across multiple genotypes.
Although the sequences of the coding and flanking regions of the HvFT1 alleles from barley varieties Morex, IMC, and BGS213 were identical, only the BGS213 allele was sufficient to induce early flowering in the presence of vrn-H1 and Vrn-H2 alleles for winter growth habit. These differences motivated the study of HvFT1 copy number in these varieties and led to the discovery of the increased HvFT1 copy number in BGS213. Surprisingly, the BGS213 allele included 4–5 copies of HvFT1, which was confirmed in the two backcross introgression lines Hayakiso 2-Tammi and Dairokkaku 1-Tammi (Fig. 1). No HvFT1 duplications were observed among the other eight varieties characterized for HvFT1 copy number in this study. This observation, together with the lack of sequence differences among the 4–5 HvFT1 copies present in the BGS213 allele, suggest that this duplication is of recent origin.
The HvFT1 allele for spring growth habit found in BGS213 comes from the spring variety Tammi (Olli/Asplund, both spring), which was released in Finland prior to 1950 (received by the National Small Grain Collection in 1949 as PI 175505). This variety can be found in the pedigrees of barley varieties from Finland, Sweden, Canada, and Alaska, where the short growing seasons require varieties with very early flowering and short life cycles.
Additional HvFT1 alleles dominant for spring growth habit have been reported in barley accessions from North Pakistan, North India, Tibet, and Ethiopia in the germplasm collection at Okayama University (Takahashi 1983). Here, we confirmed the presence of dominant spring HvFT1 alleles in one accession from Ethiopia (E878) and another one from Russia (U672), and showed that these two alleles were not associated with differences in HvFT1 copy number (both alleles showed the PE ITC haplotype combination).
With the identification in this study of CNV at the HvFT1 locus, all four major flowering genes in the Triticeae have been reported to have some level of CNV. Two to four copies of the Photoperiod-B1 gene were identified on wheat chromosome 2B and two to three copies of the vernalization gene VRN-A1 were detected recently on wheat chromosome 5A (Díaz et al. 2012). We reported before the presence of CNV of the ZCCT genes in both wheat and barley (Dubcovsky et al. 2005; Distelfeld et al. 2009). Other important agronomic genes in the Triticeae also show CNV, including a tandem segmental duplication of the wheat dwarfing gene Rht-D1 (Li et al. 2012), and duplications of the frost tolerance CBF transcription factors in barley and wheat (Knox et al. 2010). The rapid increase in CNV examples reported in barley and wheat genes likely reflects the relatively high frequency of gene copy number differences in these species. It has been proposed that the abundance of repetitive elements in the large genomes of the Triticeae species might be associated with the faster rates of duplications and deletions observed in these species (Dubcovsky and Dvorak 2007; Saintenac et al. 2011; Wicker et al. 2011).
Effect of increased copy number on gene expression and phenotype
In Arabidopsis, which has a small genome and limited numbers of repetitive sequences, copy number variation affects approximately 9 % of the genes (Gan et al. 2011). Many of these CNV are likely pseudogenes since only a small proportion is expressed. Among the 388 expressed genes with CNV, only 54 showed differences in expression that were attributed to CNV, which suggested that CNV has a modest effect on differences in gene expression in Arabidopsis. However, several examples of differential expression of genes associated with CNV and phenotypic changes have started to emerge in barley and wheat, suggesting the possibility that the faster rates of duplications (Dubcovsky and Dvorak 2007), together with the strong selection pressures imposed by agriculture may contribute to a larger role of CNV on expression and phenotypic differences in these species than in Arabidopsis.
In wheat, the increased copy number observed in the Ppd-B1 locus was associated with significantly higher transcript levels particularly at dawn when expression in the wild type was very low (Díaz et al. 2012). In contrast, the increased copy number of recessive vrn-A1 alleles was associated with slower induction of expression, which is consistent with the increased vernalization requirement and the delayed flowering time observed in the varieties with higher vrn-A1 copy number (Díaz et al. 2012). Large dosage effects of the VRN-H2 locus were observed here in the IMC x Hayakiso 2 segregating population, suggesting that the natural variation in VRN-H2 copy number should also have an effect on flowering time. Natural variation in VRN2 copy number has been previously described in wheat (Distelfeld et al. 2009). Additional CNV affecting gene expression and phenotype have been reported in genes affecting other important agronomic traits in barley and wheat (Li et al. 2012; Knox et al. 2010; Stockinger et al. 2007; Ragupathy et al. 2008). These examples show that CNV can contribute to important adaptive variation in the Triticeae species through changes in the patterns of gene expression.
Using the Hayakiso 2 × Hayakiso 2-Tammi population, we showed that CNV can influence the timing of expression. Plants homozygous for the Tammi HvFT1 allele (4–5 HvFT1 copies) had earlier HvFT1 expression during development than the near isogenic lines with a single HvFT1 gene. Heterozygous plants (average ~3 copies per genome) showed an intermediate initiation time of HvFT1 expression levels (Fig. 4) providing additional evidence of the association between HvFT1 copy number, expression levels and flowering time.
A correlation between earlier increases in HvFT1 transcript levels and earlier heading time was also observed in wheat. Recombinant substitution lines carrying the TaFT-B1 allele from the variety Hope, which has a retrotransposon insertion in the promoter showed higher FT1 transcript levels and earlier flowering time than the isogenic lines carrying the wild type TaFT-B1 allele (Yan et al. 2006). Similarly, a winter wheat line transformed with the TaFT-B1 Hope allele showed early flowering even in the absence of vernalization (Yan et al. 2006). To test if the early flowering was associated with CNV we estimated TaFT-B1 copy number in the transgenic lines relative to the non-transgenic Hope allele. Analysis of genomic DNAs from multiple plants from two independent transgenic events (Yan et al. 2006) showed that these transgenic lines have approximately four (3.7 ± 0.27, 8 plants) and five (4.70 ± 0.14, 12 plants) TaFT-B1 copies. Although these differences in copy number may explain the earlier induction of TaFT-B1 in the transgenic wheat plants, we cannot rule out alternative explanations. Since these transgenic events were produced by bombardment, some of the inserted copies may not be functional. Altered transcript levels can be also generated by the truncation of critical regulatory regions in the construct and/or rearrangements during the transformation.
Haplotype differences in HvFT1 promoter and first intron
Based on the early flowering associated with the Tammi and BGS213 HvFT1 alleles, Yan et al. (2006) hypothesized that the IAG intron one haplotype present in these varieties might be associated with their early flowering time, but cautioned that the number of varieties analyzed was insufficient to make valid conclusions. The results from the present study indicate that the early flowering associated with the HvFT1 alleles from Tammi and BGS213 described by Yan et al. (2006) is most likely the result of their increased copy number. When lines with a single HvFT1 copy are compared, the intron one IAG haplotype seems to be associated with a delay in flowering relative to the ITC haplotype. In the IMC x Hayakiso 2 population described here, plants homozygous for the IAG haplotype flowered 10–13 days later than the plants heterozygous or homozygous for the ITC haplotype (both PE promoter). Although these differences were not significant (likely due to high variability of the late flowering plants), a similar result was described recently in the cross between the French variety Esterel and the Spanish landrace SBCC016. These two varieties have identical HvFT1 promoter haplotypes (PL) but different intron one haplotypes. In this population, plants homozygous for the IAG haplotype flowered 7 days later (P < 0.01) than the plants homozygous for the ITC haplotype (Casas et al. 2011). This result was further validated in a collection of 140 winter barley landraces, in which the IAG haplotype was associated with 6–8 days later flowering than the ITC haplotype and showed a high correlation with latitude (R = 0.55; Casas et al. 2011). Cuesta-Marcos et al. (2010) also reported the presence of the IAG haplotype in a winter variety confirming that this haplotype is not associated with early flowering. The association between haplotype differences in the HvFT1 intron and flowering time is not surprising given the important role of FT intron 1 polymorphisms on flowering reported in other species (Adrian et al. 2010; Helliwell et al. 2006; Schwartz et al. 2009; Tiwari et al. 2010). However, a conclusive determination of a causal relationship between the ITC haplotype and early flowering will require experimental validation using transgenic approaches.
Results from this study also showed significant associations between HvFT1 promoter haplotypes and heading time in lines with a single copy of HvFT1, but these differences were not consistent among three populations that used the same H. vulgare ssp. spontaneum accession as one of the parental lines. Plants carrying the PE haplotype flowered earlier than those carrying the PL haplotype both in the E878 × H. vulgare ssp. spontaneum and U672 × H. vulgare ssp. spontaneum segregating populations. However, in the Hayakiso 2 × H. vulgare ssp. spontaneum population, plants homozygous for the PE haplotype flowered 10–14 days later than those carrying the PL haplotype (P < 0.0001). This unexpected result was confirmed by independent experiments in two laboratories. The same trend was observed when the differences between the HvFT1 alleles were analyzed only within the class homozygous for the VRN-H1 allele from H. vulgare ssp. spontaneum (Table 4), which was the same accession used in the E878 and U672 populations. This last result indicates that the inconsistent effect of the HvFT1 PE allele from Hayakiso 2 is not caused by epistatic effects of the VRN-H1 allele.
Taken together, the previous results suggest that the observed differences in the promoter haplotypes are not the cause of the differences in flowering time, but are just markers linked to a yet unknown cause of these differences. The heterogeneous effects of the promoter haplotypes may also explain the limited effect of the promoter haplotypes observed in the study of 140 Spanish winter barley landraces (SBCC; Casas et al. 2011). In this collection, the PE haplotype was associated with 2–3 days earlier flowering in the fall sowing experiment, but with an opposite effect in the April sowing experiment (3.2 days later flowering within the IAG lines). In a subsequent doubled haploid population from the cross SBCC145 (PE ITC) x Beatrix (PL ITC), the PE haplotype was associated with an earlier heading time, though the differences were significant only within the photoperiod insensitive class (Ponce-Molina et al. 2012). In our E878 (PE ITC) × H. vulgare ssp. spontaneum (PL ITC) segregating population, the effects of HvFT1 on heading time was significant both in photoperiod sensitive and insensitive backgrounds.
Epistatic interactions between VRN-H1 and HvFT1 alleles
The heterogeneity of the effects of the promoter haplotypes on heading time was also evident in their epistatic effects. In the E878 × H. vulgare ssp. spontaneum and U672 × H. vulgare ssp. spontaneum populations, the plants carrying the HvFT1 PE ITC haplotype showed a spring growth habit irrespectively of the VRN-H1 allele. This result indicates that this HvFT1 allele is epistatic to the VRN-H1 and VRN-H2 alleles for winter growth habit present in these populations (Fig. 5). In spite of having an identical HvFT1 haplotype (PE ITC), the Hayakiso 2 allele was not able to overcome the vernalization requirement in the Hayakiso 2 × H. vulgare ssp. spontaneum population (Table 4). Based on this result, we conclude that the currently known differences in the sequences of the HvFT1 promoter haplotypes are not sufficient to explain the different epistatic interactions involving this locus.
The increased HvFT1 copy number found in the BGS213 and Tammi alleles was associated with stronger and more consistent epistatic interactions than the haplotype differences among single copy alleles described above. In the BGS213 × H. vulgare ssp. spontaneum and BGS213 × Igri populations, the early flowering HvFT1 allele from BGS213 was epistatic to the alleles for winter growth habit (Yan et al. 2006). Similar epistatic interactions and early flowering were observed when the HvFT1 allele with increased copy number was introgressed into the varieties Hayakiso 2 and Dairokkaku 1, which have a strong vernalization requirement (Yan et al. 2006), and in the early experiments using the barley variety Tammi (Takahashi and Yasuda 1971). These results indicate that epistatic effects of the HvFT1 allele from BGS213/Tammi are consistent across different genotypes.
Conclusions and practical applications
From a practical point of view, our results have clarified the effects of HvFT1 natural variation on barley flowering time. We demonstrated that the BGS213 genetic stock used to define the dominant Vrn-H3 spring allele includes multiple copies of HvFT1 that result in earlier expression and earlier flowering time. The large effect of this allele on flowering time may restrict its use to areas or cropping systems that require short growing cycles. In contrast, the allelic differences in HvFT1 first intron are associated with smaller effects on flowering time that can be used to fine tune flowering time of barley varieties to different or changing environments. The presence of multiple HvFT1 alleles with diverse effects on flowering time suggests that natural variation at this locus may have contributed to the wide adaptation of barley to different environments.
From a more basic point of view, our results add to a rapidly growing literature on CNV in the Triticeae species. CNV has been found for most of the flowering genes studied so far in the temperate grasses, suggesting that this mechanism plays an important role in the generation of novel diversity. This is not surprising given the dynamic nature of the large genomes of the Triticeae species that exhibit rates of duplications and deletions several orders of magnitude faster than the rates of nucleotide substitution (Dubcovsky and Dvorak 2007; Saintenac et al. 2011; Wicker et al. 2011). In the long term, gene duplications play an important evolutionary role, as they provide opportunities for diversification and sub-functionalization that can increase adaptative plasticity.
Acknowledgments
This project was supported by the National Research Initiative Competitive Grants No. 2011-67013-30077 and No. 2011-68002-30029 (Triticeae-CAP) from the USDA National Institute of Food and Agriculture and by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation.
References
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F. cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell. 2010;22:1425–1440. doi: 10.1105/tpc.110.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, Drosse B, Searle I, Coupland G, von Korff M. Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J. 2012;69:868–880. doi: 10.1111/j.1365-313X.2011.04839.x. [DOI] [PubMed] [Google Scholar]
- Casao MC, Karsai I, Igartua E, Gracia MP, Veisz O, Casas AM. Adaptation of barley to mild winters: a role for PPD-H2. BMC Plant Biol. 2011;11:164. doi: 10.1186/1471-2229-11-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas AM, Djemel A, Ciudad FJ, Yahiaoui S, Ponce LJ, Contreras-Moreira B, Gracia MP, Lasa JM, Igartua E. HvFT1 (VrnH3) drives latitudinal adaptation in Spanish barleys. Theor Appl Genet. 2011;122:1293–1304. doi: 10.1007/s00122-011-1531-x. [DOI] [PubMed] [Google Scholar]
- Chen A, Dubcovsky J. Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 2012;8:e1003134. doi: 10.1371/journal.pgen.1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Coupland G. Photoperiodic flowering of Arabidopsis: integrating genetic and physiological approaches to characterization of the floral stimulus. Plant Cell Environ. 2005;28:54–66. doi: 10.1111/j.1365-3040.2005.01283.x. [DOI] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Cuesta-Marcos A, Szucs P, Close TJ, Filichkin T, Muehlbauer GJ, Smith KP, Hayes PM. Genome-wide SNPs and re-sequencing of growth habit and inflorescence genes in barley: implications for association mapping in germplasm arrays varying in size and structure. BMC Genomics. 2010;11:707. doi: 10.1186/1471-2164-11-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 2003;132:1849–1860. doi: 10.1104/pp.103.023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum) PLoS ONE. 2012;7:e33234. doi: 10.1371/journal.pone.0033234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Tranquilli G, Li C, Yan L, Dubcovsky J. Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol. 2009;149:245–257. doi: 10.1104/pp.108.129353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Chen CL, Yan LL. Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Mol Breed. 2005;15:395–407. doi: 10.1007/s11032-005-0084-6. [DOI] [Google Scholar]
- Faure S, Higgins J, Turner A, Laurie DA. The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare) Genetics. 2007;176:599–609. doi: 10.1534/genetics.106.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franckowiak JD, Konishi T. Barley Genetic Stock 213: spring growth habit 3. Barley Genet Newsl. 1997;26:212. [Google Scholar]
- Fu D, Szücs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, Kahles A, Bohnert R, Jean G, Derwent P, Kersey P, Belfield EJ, Harberd NP, Kemen E, Toomajian C, Kover PX, Clark RM, Raetsch G, Mott R. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006;46:183–192. doi: 10.1111/j.1365-313X.2006.02686.x. [DOI] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B. Low-temperature and day length cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol. 2008;147:355–366. doi: 10.1104/pp.108.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming MN, Fieg S, Peacock WJ, Dennis ES, Trevaskis B. Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol Genet Genomics. 2009;282:107–117. doi: 10.1007/s00438-009-0449-3. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Kawahigashi H, Ando T, Tonooka T, Handa H. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 2009;149:1341–1353. doi: 10.1104/pp.108.132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox AK, Dhillon T, Cheng H, Tondelli A, Pecchioni N, Stockinger EJ. CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet. 2010;121:21–35. doi: 10.1007/s00122-010-1288-7. [DOI] [PubMed] [Google Scholar]
- Li C, Dubcovsky J. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 2008;55:543–554. doi: 10.1111/j.1365-313X.2008.03526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Distelfeld A, Comis A, Dubcovsky J. Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J. 2011;67:763–773. doi: 10.1111/j.1365-313X.2011.04630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xiao J, Wu J, Duan J, Liu Y, Ye X, Zhang X, Guo X, Gu Y, Zhang L, Jia J, Kong X. A tandem segmental duplication (TSD) in green revolution gene Rht-D1b region underlies plant height variation. New Phytol. 2012;196:282–291. doi: 10.1111/j.1469-8137.2012.04243.x. [DOI] [PubMed] [Google Scholar]
- Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J. Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol. 2005;138:2364–2373. doi: 10.1104/pp.105.064287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SN, Finnegan EJ, Dennis ES, Peacock WJ, Trevaskis B. Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc Natl Acad Sci USA. 2009;106:8386–8391. doi: 10.1073/pnas.0903566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan A, Hayes PM, Chen F, Chen THH, Blake T, Wright S, Karsai I, Bedo Z. Genetic-analysis of the components of winterhardiness in barley (Hordeum vulgare L.) Theor Appl Genet. 1994;89:900–910. doi: 10.1007/BF00224516. [DOI] [PubMed] [Google Scholar]
- Ponce-Molina LJ, María Casas A, Pilar Gracia M, Silvar C, Mansour E, Thomas WBT, Schweizer G, Herz M, Igartua E. Quantitative trait loci and candidate loci for heading date in a large population of a wide barley cross. Crop Sci. 2012;52:2469–2480. doi: 10.2135/cropsci2012.01.0029. [DOI] [Google Scholar]
- Ragupathy R, Naeem HA, Reimer E, Lukow OM, Sapirstein HD, Cloutier S. Evolutionary origin of the segmental duplication encompassing the wheat GLU-B1 locus encoding the overexpressed Bx7 (Bx7OE) high molecular weight glutenin subunit. Theor Appl Genet. 2008;116:283–296. doi: 10.1007/s00122-007-0666-2. [DOI] [PubMed] [Google Scholar]
- Read BJ, Raman H, McMichael G, Chalmers KJ, Ablett GA, Platz GJ, Raman R, Genger RK, Boyd WJR, Li CD, Grime CR, Park RF, Wallwork H, Prangnell R, Lance RCM. Mapping and QTL analysis of the barley population Sloop × Halcyon. Aust J Agric Res. 2003;54:1145–1153. doi: 10.1071/AR03037. [DOI] [Google Scholar]
- Saintenac C, Jiang D, Akhunov ED. Targeted analysis of nucleotide and copy number variation by exon capture in allotetraploid wheat genome. Genome Biol. 2011;12:R88. doi: 10.1186/gb-2011-12-9-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasani S, Hemming MN, Oliver SN, Greenup A, Tavakkol-Afshari R, Mahfoozi S, Poustini K, Sharifi HR, Dennis ES, Peacock WJ, Trevaskis B. The influence of vernalization and day length on expression of flowering-time genes in the shoot apex and leaves of barley (Hordeum vulgare) J Exp Bot. 2009;60:2169–2178. doi: 10.1093/jxb/erp098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, Balasubramanian S, Warthmann N, Michael TP, Lempe J, Sureshkumar S, Kobayashi Y, Maloof JN, Borevitz JO, Chory J, Weigel D. Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana. Genetics. 2009;183:723–732. doi: 10.1534/genetics.109.104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Skinner JS, Gardner KG, Francia E, Pecchioni N. Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J. 2007;51:308–321. doi: 10.1111/j.1365-313X.2007.0141.x. [DOI] [PubMed] [Google Scholar]
- Takahashi R. Catalogue of the barley germplasm preserved in Okayama University. Institute of Agricultural and Biological Sciences. Kurashiki: Okayama University; 1983. p. 217. [Google Scholar]
- Takahashi R, Yasuda S. Genetics of earliness and growth habit in barley. In: Nilan RA, editor. Proceedings of the 2nd International Barley Genetics Symposium. Washington: Washington State University Press; 1971. pp. 388–408. [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima C, Shimamoto K. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang H-C, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, Belachew A, Repetti PP, Reuber TL, Ratcliffe OJ. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010;187:57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA. 2003;100:13099–13104. doi: 10.1073/pnas.1635053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES. HvVRN2 responds to day length, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 2006;140:1397–1405. doi: 10.1104/pp.105.073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007;12:352–357. doi: 10.1016/j.tplants.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Tsuji H, K-i Taoka, Shimamoto K. Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr Opin Plant Biol. 2011;14:45–52. doi: 10.1016/j.pbi.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- von Zitzewitz J, Szücs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen TH, Hayes PM, Skinner JS. Molecular and structural characterization of barley vernalization genes. Plant Mol Biol. 2005;59:449–467. doi: 10.1007/s11103-005-0351-2. [DOI] [PubMed] [Google Scholar]
- Weaver S, Dube S, Mir A, Qin J, Sun G, Ramakrishnan R, Jones RC, Livak KJ. Taking qPCR to a higher level: analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods. 2010;50:271–276. doi: 10.1016/j.ymeth.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Wicker T, Mayer KF, Gundlach H, Martis M, Steuernagel B, Scholz U, Šimková H, Kubaláková M, Choulet F, Taudien S, Platzer M, Feuillet C, Fahima T, Budak H, Doležel J, Keller B, Stein N. Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell. 2011;23:1706–1718. doi: 10.1105/tpc.111.086629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Yan L, Echenique V, Busso C, SanMiguel P, Ramakrishna W, Bennetzen JL, Harrington S, Dubcovsky J. Cereal genes similar to Snf2 define a new subfamily that includes human and mouse genes. Mol Genet Genomics. 2002;268:488–499. doi: 10.1007/s00438-002-0765-3. [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, von Zitzewitz J, Skinner JS, Hayes PM, Dubcovsky J. Molecular characterization of the duplicated meristem identity genes HvAP1a and HvAP1b in barley. Genome. 2005;48:905–912. doi: 10.1139/g05-035. [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]


