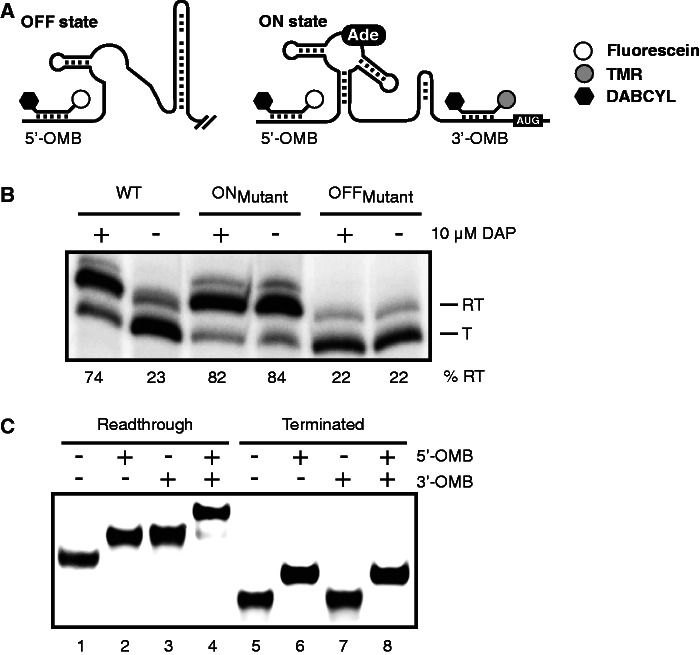
Figure 1.
Strategy for monitoring riboswitch activity using molecular beacons. (A) Schematic representation of the pbuE riboswitch regulation, including hybridization sites for both molecular beacons. In absence of adenine, the B. subtilis pbuE riboswitch folds in the OFF state and forms a terminator stem that prematurely stops transcription. However, on adenine (Ade) binding, the terminator stem is destabilized, and transcription elongation is allowed (ON state). Although the 5′ molecular beacon (5′-OMB) can hybridize to both OFF and ON state transcripts, the 3′ beacon (3′-OMB) is only able to bind to the elongated ON state transcript. Both molecular beacons contain a quencher (DABCYL, black hexagon) at the 3′ extremity, preventing fluorescence emission when beacons are not hybridized to the riboswitch. TMR and fluorescein are represented by gray and white circles, respectively. (B) Single-round in vitro transcription assays performed in absence (−) and in presence (+) of 10 µM DAP for the wild-type (WT), ON and OFF state mutants. Readthrough (RT) and terminated (T) transcripts are shown on the right of the gel. Percentages of termination are indicated later in the text the gel. (C) EMSA performed on the pbuE riboswitch elongated (lanes 1–4) and terminated (lanes 5–8) transcripts in complex with indicated molecular beacons. Free and bound riboswitch transcripts were separated on polyacrylamide gel in native conditions.