Figure 3.
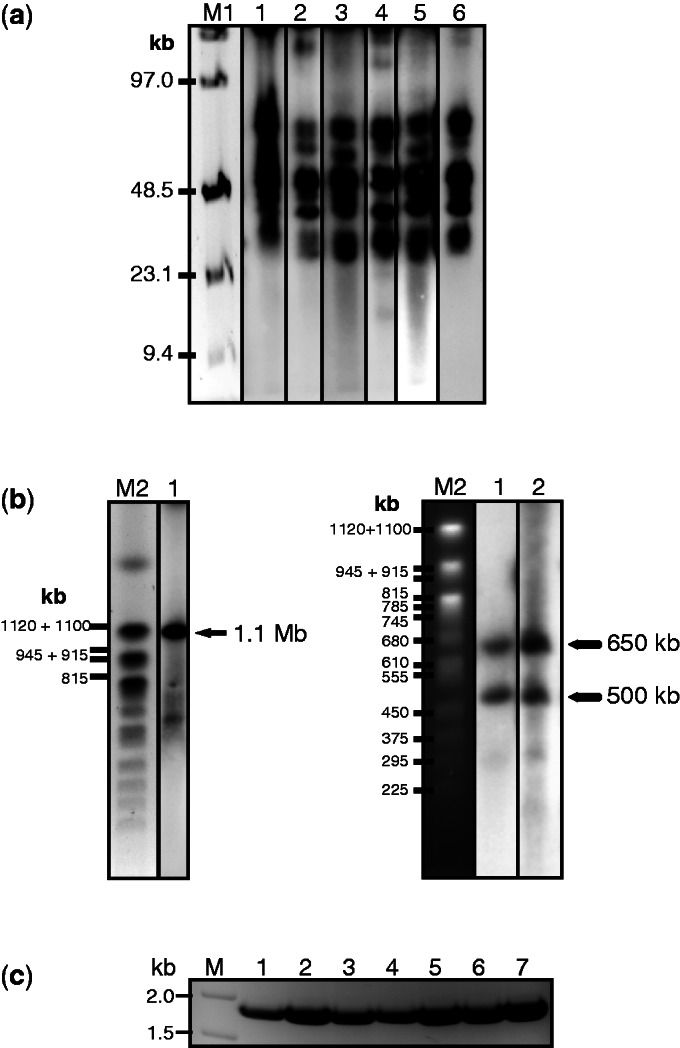
Physical analysis of the alphoidtetO-HAC used for loading of tTS-containing cassettes. (a) Analysis of integrity of the alphoidtetO-HAC synthetic array after its transfer into HPRT-deficient HT1080 cells. Genomic DNA from the cells with the HAC was digested with SpeI endonuclease, separated by CHEF gel electrophoresis (range 10–70 kb) and the transferred membrane was hybridized with the tetO-alphoid probe. Lane 1: the HAC with the loxP site in hamster CHO cells; lanes 2, 3, 4 and 5: four HAC clones with the loxP site in HPRT-deficient HT1080 cells; lane 6: the original HAC (clone AB2.218.21) generated in human cells (29). M1- Pulse Marker™ 0.1–200 kb (Sigma-Aldrich). (b) Mapping of the loxP site in a mega-base size alphoid DNA array. Genomic DNA from the cells possessing the original HAC was digested with PmeI endonuclease, separated by CHEF gel electrophoresis (range 200–1500 kb) and the transferred membrane was hybridized with the tetO-alphoid probe. A single 1.1-Mb fragment was detected for the original HAC (left, lane 1). Two bands of 650 and 500 kb were detected for genomic DNA from cells with the HAC bearing an inserted NBS1 gene (right, lanes 1 and 2). The size of the fragments was determined by comparison with the DNA size standard, Saccharomyces cerevisiae chromosomes. Lane M2: Yeast Chromosome PFG Marker BioLabs. (c) PCR analysis of clones with insertion of X3.1-I-EGFP-I into the HAC confirming restoration of the full-length HPRT gene.
