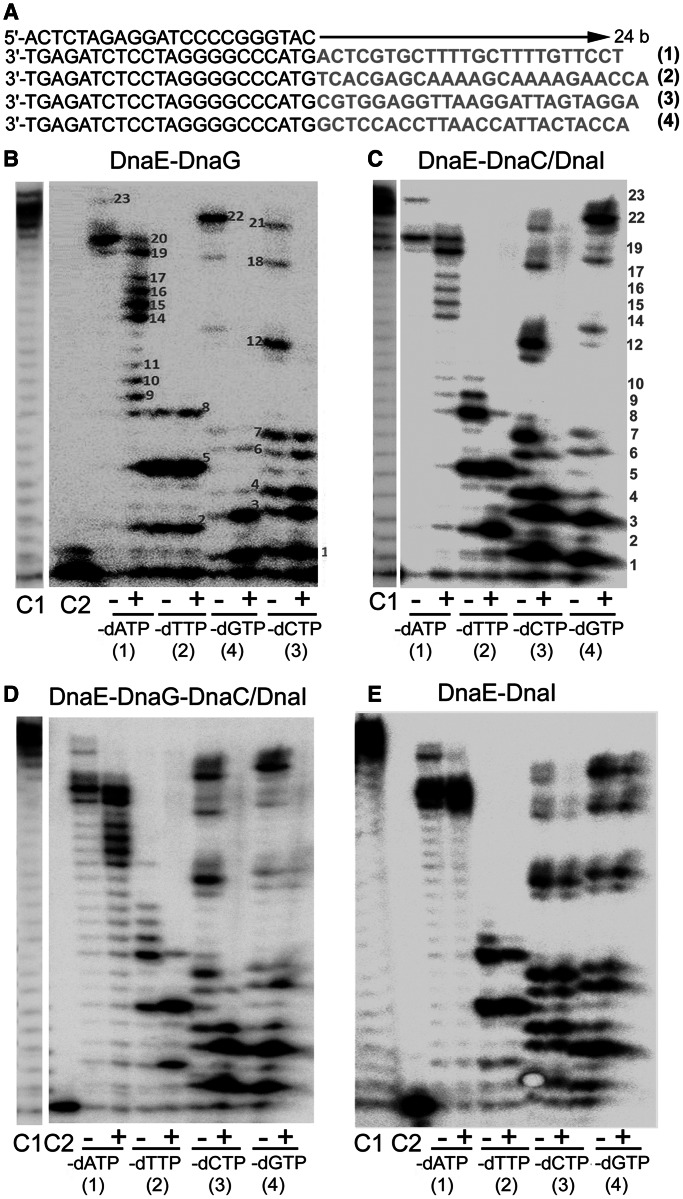
Figure 10.
Effects of the DnaG primase and DnaC/DnaI helicase on the error-prone DnaEBs polymerase activity. (A) The sequences of the oligonucleotides used to prepare the primer extension substrates are shown. Each of the bottom four oligonucleotides was annealed to the top oligonucleotide to produce substrates 1–4. The sequences shown in pale shading were the templates used by the DnaEBs polymerase during primer extensions with these substrates. (B–E) Representative urea–polyacrylamide denaturing gels showing the effects of DnaG (30 nM) (B), DnaC/DnaI (60 nM each protein) (C), DnaG+DnaC/DnaI (D) and DnaI (60 nM) (E) on the error-prone polymerase activity of DnaEBs (10 nM). Lanes labelled C1 and C2 represent primer extension reactions with (showing full extension of the DNA substrate) and without (showing no extension) DnaEBs, respectively, in the presence of all four dNTPs. Lanes labelled with − and + signs represent DnaEBs extension reactions in the absence and presence of the relevant effector proteins in the different substrates (1–4) shown underneath the gels. The missing dNTP nucleotide for each reaction is indicated underneath each gel. All reactions were carried out at 37°C for 16 min.