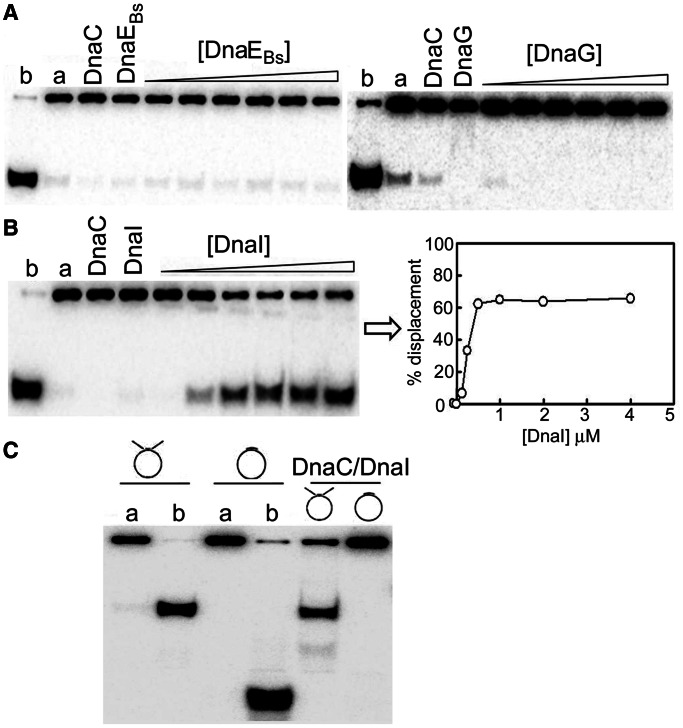
Figure 5.
Characterization of the B. subtilis DnaC helicase activity. (A) The DnaC helicase activity was assayed by monitoring the displacement of a 104-mer oligonucleotide annealed onto ssM13 and forming a double-fork substrate with poly(dT) tails. DnaC did not exhibit any detectable helicase activity on its own or in the presence of the DnaG primase and the DnaEBs polymerase. Control reactions with DnaG and DnaEBs confirmed, as expected, that these proteins have no helicase activities. Control samples in lanes a and b represent annealed substrate and fully displaced (boiled) controls. Reactions were carried out with 500 nM monomeric DnaC incubated for 1 h at 37°C as described in ‘Materials and Methods’ section, with increasing concentrations of DnaE (40–1300 nM) or DnaG (125–4000 nM). (B) The DnaC helicase activity was detectable only in the presence of the putative DnaI helicase loader. Similar helicase reactions were carried out as above in the presence of DnaI (125, 250, 500, 1000, 2000–4000 nM monomeric). Data were plotted as a percentage of the radiolabelled oligonucleotide displacement versus DnaI concentration. Maximal stimulation of the DnaC helicase activity was observed at 500 nM DnaI, indicating optimal 1:1 stoichiometry consistent with a DnaC6:DnaI6 complex. (C) The helicase activity requires a fork DNA substrate. Helicase reactions were carried out as above for 20 min at 37°C with DnaC (500 nM) and DnaI (500 nM) in the presence or absence of DnaG (250 nM) with two different DNA substrates (with or without fork), as indicated. The double-fork substrate forms 5′ and 3′ poly(dT) tails, and the non-fork DNA substrate was the same used in the polymerase assay with a synthetic oligonucleotide annealed onto ssM13 (see ‘Materials and Methods’ section). Control samples in lanes a and b represent annealed substrate and fully displaced (boiled) controls.