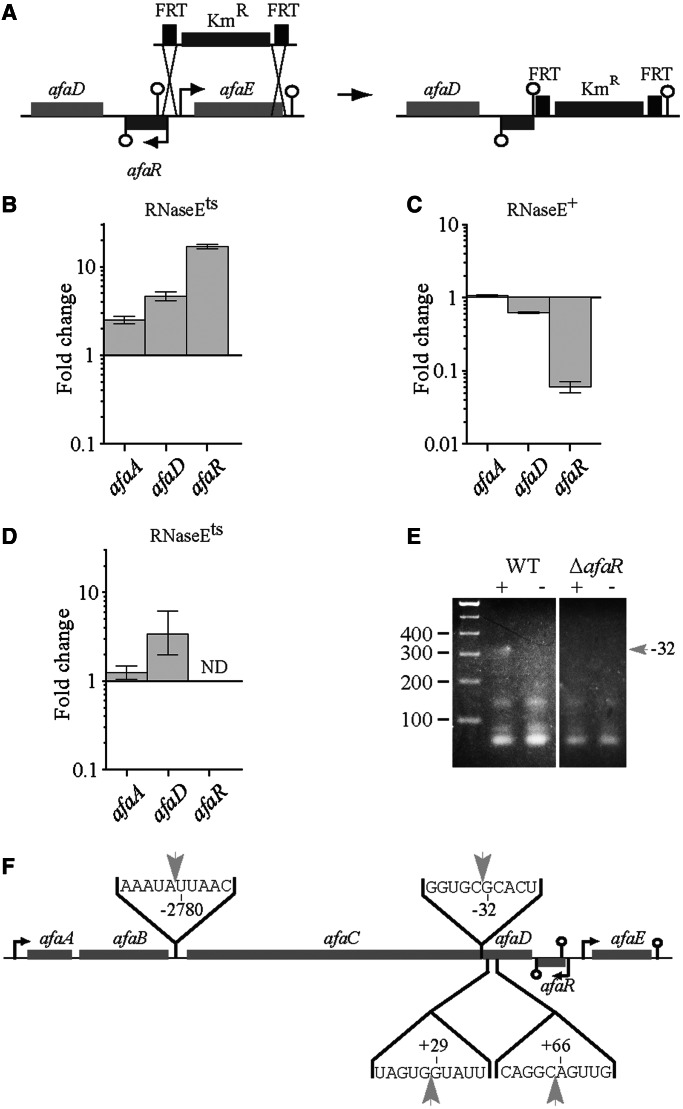
Figure 6.
RNase E dependence of the regulation of afaD expression by AfaR. (A) Schematic representation of the construction of the pILL1323 plasmid by allelic exchange from pILL1320. qRT-PCR analysis confirmed an absence of afaR expression from pILL1323 (See Supplementary Figure S1). (B–D) Relative levels of the afaA and afaD mRNAs and AfaR sRNA, determined by qRT-PCR, in wild-type and RNase Ets thermosensitive strains. The values for each gene are expressed as a ratio of expression at 42°C (low level of RNase E) to that at 30°C (high level of RNase E) for the N3431 + pILL1320 strain (B, [RNase Ets]), the N3433 + pILL1320 strain (C, [RNaseE+]) and the N3431 + pILL1322 strain (D, [RNase Ets]). (E) Circular RACE mapping of the afaC mRNA ends. Total RNA from E. coli BW25113 carrying pILL1320 (wild type) or pILL1323 (ΔafaR) was circularized by end-ligation with or without T4 RNA ligase (lanes + and −, respectively). The ligated 5′ and 3′ ends of the fragment were then amplified by RT-PCR. PCR products were analysed by electrophoresis in a 3% agarose gel. The band of interest was excised, cloned and sequenced. A ∼310 bp DNA fragment (gray arrow) was more abundant after ligation treatment (lane +) in the wild-type strain than in the ΔafaR strain, indicating an AfaR-dependent amplification of the 5′ (cleavage at position -32) and 3′ ends of the afaC mRNA. This suggests that AfaR is involved in the RNase-dependent cleavage of the afaABCD mRNA. (F) Location of RNase cleavage sites in the afaABCD mRNA.