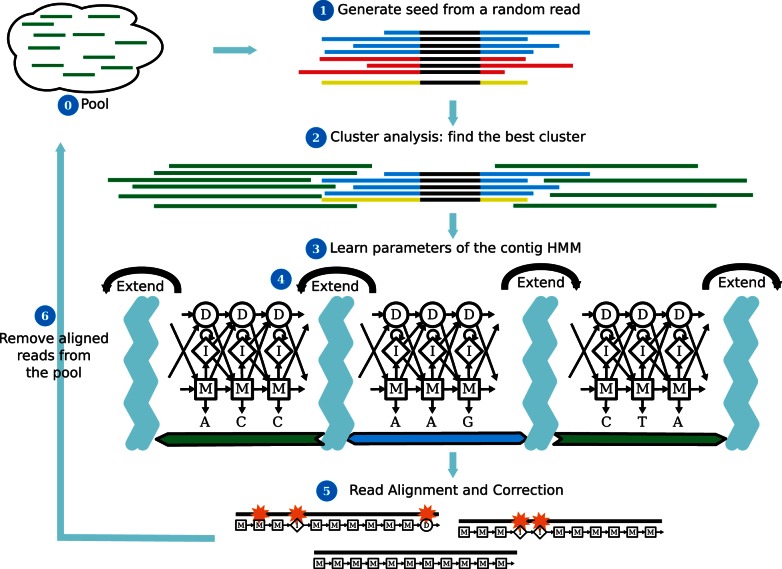
Figure 1.
An overview of SEECER. Step 1: We select a random read that has not yet been assigned to any contig HMM. Next, we extract all reads with at least k consecutive nucleotides that overlap with the selected read. Step 2: We cluster all reads and then select the most coherent subset as the initial set of the contig HMM. Step 3: We learn an initial HMM using the alignment specified by the k-mer matches of selected reads. Step 4: We use the consensus sequence defined by the contig HMM to extract additional reads from our unassigned set. These additional reads are used to extend the HMM in both directions. Step 5: When no more reads can be found to extend the HMM, we determine for each of the reads that were used to construct the HMM the likelihood of being generated by this contig HMM. For those with a likelihood above a certain threshold, we use the HMM consensus to correct errors. Step 6: We remove the reads that are assigned or corrected from the unassigned pool. See ‘Materials and Methods’ section for complete details.