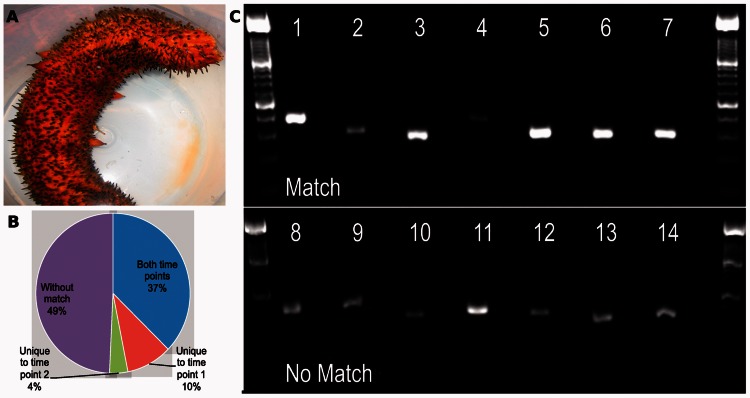
Figure 4.
De novo assembly of sea cucumber data. (A) A living sea cucumber Parastichopus parvimensis. (B) Distribution of BlastX matches of sea cucumber transfrags to known sea urchin peptides. The percentages represent the subset of sea urchin peptides that we have significantly matched to at least one transfrag in time point 1 and/or time point 2 and those that were not matched to any transfrag. (C) Ethidium bromide–stained image of PCR products amplified from sea cucumber cDNA. Primer pairs were designed against 14 assembled transfrags, seven of which matched to known peptides of RNAs (top row), and seven other that had no match in the database (bottom row). Standard ladders of 100-bp size are in the first and last lanes. Each lane is followed by the appropriate no template control to demonstrate that amplification was not due to non-specific contamination.