Abstract
We present an intramolecular reaction, Reflex™, to derive shorter, sequencer-ready, daughter polymerase chain reaction products from a pooled population of barcoded long-range polymerase chain reaction products, whilst still preserving the cognate DNA barcodes. Our Reflex workflow needs only a small number of primer extension steps to rapidly enable uniform sequence coverage of long contiguous sequence targets in large numbers of samples at low cost on desktop next-generation sequencers.
INTRODUCTION
Current next-generation sequencing (NGS) platforms require that adaptors are appended to the ends of target DNA sequences for use in the sequencing reaction. An extra multiplex identifier (MID), a short DNA barcode that identifies the sample, is added with the sequencing adaptor to sequence multiple DNA samples in a single sequencing run. Typically, individual samples are prepared and pooled immediately before sequencing, requiring expensive and labour-intensive preparation methods: thus, for targeted re-sequencing, sample preparation costs dominate the overall cost.
For example, for a large population of DNA samples, to obtain sequence coverage of a long contiguous region that exceeds the read length of the NGS platform, enrichment of a genomic sequence target is achieved by long-range polymerase chain reaction (LRPCR). The next step is to derive shorter fragments suitable for NGS. LRPCR products from each sample are either randomly physically fragmented followed by ligation of MIDs (1,2) or by in vitro transposition-mediated fragmentation coupled with tagging (3). Then samples are pooled for sequencing. In both of these fragmentation/tagging approaches, sample preparation costs are high, and for physical fragmentation, the resulting sequencing coverage can be variable across the target.
We were therefore motivated to create a technique that can perform sample preparation on a pooled population of long ‘parent’ DNA fragments, already appended with adaptors and MIDs, to generate smaller, sequencer-ready, ‘daughter’ amplicons—yet preserving cognate MIDs. This is done by first deriving progenitor molecules carrying short inverted repeats of a ‘Reflex’ sequence; then, intramolecular pairing of the Reflex repeats, followed by polymerase extension, copies the cognate MID at the other end of the molecule. We therefore call this the Reflex reaction after ‘reflexive’ or ‘directed back on itself’.
When combined with LRPCR for simultaneous target enrichment and sample labelling, the Reflex reaction can be used to sequence target regions in large pooled populations of samples, as we show here. However, it can also be applied in other situations where there is a need to create smaller molecules from a labelled population of larger molecules.
MATERIALS AND METHODS
We obtained 95 genomic DNA samples from the Coriell Institute for Medical Research. All samples originated from the CEPH collection of the International HapMap project and were made up of 26 trios, 5 duos and 2 singletons, together with 5 duplicated samples. Primers were designed using Primer3 with default parameters.
To generate MID-tagged CYP2D6 amplicons, we set-up one 50 µl LRPCR per sample plus one water control in a 96-well plate (primer sequences are provided in Supplementary Table S3). The reactions contained 1× GoTaq Reaction Buffer (Promega), 2.5 mM MgCl2, 200 mM each dNTP, 0.4 µM each primer, 3% DMSO and 1.25 U of GoTaq Hot Start polymerase (Promega) with 250 ng of genomic DNA. We denatured the samples at 95°C for 5 min, and then ran 35 PCR cycles of 95°C for 30 s, 62°C for 30 s and 68°C for 7 min.
We checked amplification success by running 3 µl of each PCR on a 1% agarose gel, estimated the relative concentration of the products with ImageQuant (GE Healthcare) software and pooled the amplicons equimolarly (we assumed that the relative concentration of the water control was the mean of the 95 samples). We purified 300 µl of the amplicon pool with Agencourt Ampure XP beads (Beckman Coulter) and eluted into 50 µl of water. We used the Qubit dsDNA BR assay kit (Life Technologies) to quantify the pool in triplicate and diluted to a total amplicon concentration of 3 pM. This pool served as our template for the Reflex extension reactions for both the 454 Junior and Ion Torrent sequencing experiments.
We set-up the Reflex extensions in 25-µl reactions consisting of 1× Herculase II Reaction Buffer (Agilent), 0.4 mM each dNTP, 0.25 µM reflex extension primer (sequences are in Supplementary Tables S4 and S5), 1% DMSO, 0.5 µl of Herculase II Fusion DNA Polymerase (Agilent), 1.25 U of GoTaq Hot Start DNA Polymerase (Promega) and 0.3 pM pooled LRPCR products. We ran the reactions at 95°C for 5 min, 56°C for 1 min and 68°C for 10 min and then purified them with Agencourt AMPure XP beads into 20 µl of water.
To reverse the polarity of the extension products, perform the Reflex reactions and make the products double stranded, we used 15.5 µl of the purified extension reactions to set-up 25-µl reactions consisting of 1× Herculase II Reaction Buffer, 0.4 mM dNTPs, 1% DMSO, 0.2 µM 454A primer (5′-CCATCTCATCCCTGC GTGTCTCCGACTCAG-3′), 0.5 µl of Herculase II Fusion DNA Polymerase and 0.25 U of GoTaq Hot Start DNA Polymerase. We cycled the reactions at 95°C for 5 min, 56°C for 30 s, 68°C for 10 min, 95°C for 30 s, 51°C for 5 min, 68°C for 10 min, 95°C for 30 s, 56°C for 30 s and 68°C for 10 min then purified each reaction into 15 µl of water using AMPure beads as before.
To generate sequenceable amplicons, we set-up 50-µl PCRs consisting of 1× GoTaq Reaction Buffer, 2.5 mM MgCl2, 0.2 mM dNTPs, 0.25 µM 454A primer, 0.25 µM Reflex PCR primer (sequences are in Supplementary Tables S6 and S7), 1.25 U GoTaq Hot Start DNA polymerase and 10 µl of purified Reflex reaction products. We denatured the reactions at 95°C for 5 min then ran 30 cycles of 95°C for 1 min, 62°C for 1 min and 72°C for 1 min, followed by a final extension step of 72°C for 10 min.
We checked 6 µl of each reaction on a MultiNA Microchip Electrophoresis System (Shimadzu) and used the resulting concentration estimates to pool the amplicons equimolarly before sending them to an external supplier for sequencing on the 454 Junior (Titanium Amplicon) or Ion Torrent (316 chip) platforms.
Reads were trimmed for multiplex identifiers and Reflex adaptors using cutadapt (http://code.google.com/p/cutadapt/). SSAHA2 was used to align reads to the human reference genome, build 36 to produce sam files (http://www.sanger.ac.uk/resources/software/ssaha2/). Reads were primer trimmed using amptools (https://github.com/PopulationGenetics/amptools). Base calling and single-nucleotide polymorphism (SNP) calling were carried out using GATK 1.6.6 using ‘–pcr_error_rate=0.05 –deletions 0.15’ to produce VCF files.
To calculate sequencing metrics, pysam (http://code.google.com/p/pysam/) was used to extract per base sequencing depths across the target, which yielded average sequencing depths and per cent of bases within 2-fold of the mean. We used pyvcf (http://pypi.python.org/pypi/PyVCF/0.6.0) to extract number of called bases and number of called based above Q30 from the VCF files.
To calculate data for coverage plots, the negative control sample reads were first removed from the aligned sam files before analysis. Mean read depth per base and proportion of cohort at base position were both calculated and plotted using R from aligned sam files. Proportion of cohort calculation based on a minimum depth of 20 reads per sample at each base position. Gene data for plot from UCSC, CYP2D6 transcript ID:uc003bcf.
For all the duplicate samples, we calculated the ‘duplicate call consistency’ as the number of identical calls between duplicates divided by the total number of calls for the duplicates. ‘Duplicate variant consistency’ was the same, except we only considered sites where either or both samples were variants.
To calculate the 1000 Genomes concordance, we downloaded the 20110521 release (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20110521/) of the 1000 Genomes calls and lifted over from hg37 to hg36. For sites called by 1000 Genomes and by us at greater than Q30, we calculated concordance as the number of identical calls divided by the total number of calls. This gave initial discordance rates of 17/990 for 454 and 34/885 for Ion Torrent. Fourteen calls were shared in the discordant sets, and 10 of the Ion Torrent sites were low quality and assumed that 1000 Genomes is correct, giving 27 (14 shared + 3 454 + 10 Ion Torrent) discordant calls for Sanger validation. The 14 calls in both discordant sets were confirmed by Sanger sequencing as correct in our data, the others were confirmed as agreeing with 1000 Genomes (Supplementary Data).
To validate variant calls that were discordant with the 1000 Genomes project, we amplified the relevant samples by LRPCR as before, then removed unincorporated primers and dNTPs by mixing 10 µl of each reaction with 2 µl of ExoSAP-IT (USB Corporation) and incubating at 37°C for 30 min before inactivating the enzymes at 80°C for 20 min. We diluted the reaction products 10-fold and amplified shorter regions within each LRPCR amplicon using M13-tailed primers designed using Primer3. Amplifications were set-up in 50-µl volumes containing 1× GoTaq Reaction Buffer, 2.5 mM MgCl2, 200 mM each dNTP, 0.4 mM each primer and 1.25 U GoTaq Hot Start Polymerase. The thermal cycling conditions were 95°C for 2 min followed by 20 cycles of 95°C for 30 s, 56°C for 30 s and 72°C for 1 min. We checked amplification success by running 2 µl of each reaction on a 2% agarose gel and submitted the products to an external supplier for Sanger sequencing. We also set-up no-template controls for each target and confirmed that these did not return readable sequence traces.
Novel variants were filtered for amplicon bias and error bias using amptools. This demands that novel variants are seen on at least two amplicons before they are considered valid.
Cost per sample calculations were performed using (A + L + Rp + Rr)/S, where A is the adaptor cost, L is the LRPCR cost, Rp is the Reflex primer cost, Rr is the Reflex reagent cost and S is the total number of samples. A = (96 × C × 46.6), where 96 is the number of samples per Reflex population, C is the number of contiguous regions and $46.6 is the cost of adaptor oligonucleotide synthesis. L = (C × S × 1.33), where C and S are as before, and $1.33 is the cost of LRPCR reagents for each sample. Rp = [C × (Lbp/Rbp) × 21], where C is as before, Lbp = size of LRPCR in base pairs, Rbp = Lbp/(number of Reflex amplicons), set at 222 bp for 454, 63 bp for Ion Torrent, and 87 bp for Illumina, and $21 is the cost of a Reflex primer pair. Note that Rbp values for 454 and Ion Torrent are as presented here as for CYP2D6 design, whereas Rbp for Illumina is estimated using usable paired-end read lengths of 130 bp with a tiling overlap of three amplicons per target base pair. Rr = C × (S/96) × (Lbp/Rbp) × 6.45, where C, S, Lbp and Rbp are as before, 96 is the number of samples per Reflex population pool rounded up and $6.45 is the reagent cost of a Reflex reaction. Simulations with variable S and C are shown in Supplementary Table S2 for the three NGS platforms. Costs are in US dollars ($).
RESULTS
The Reflex workflow
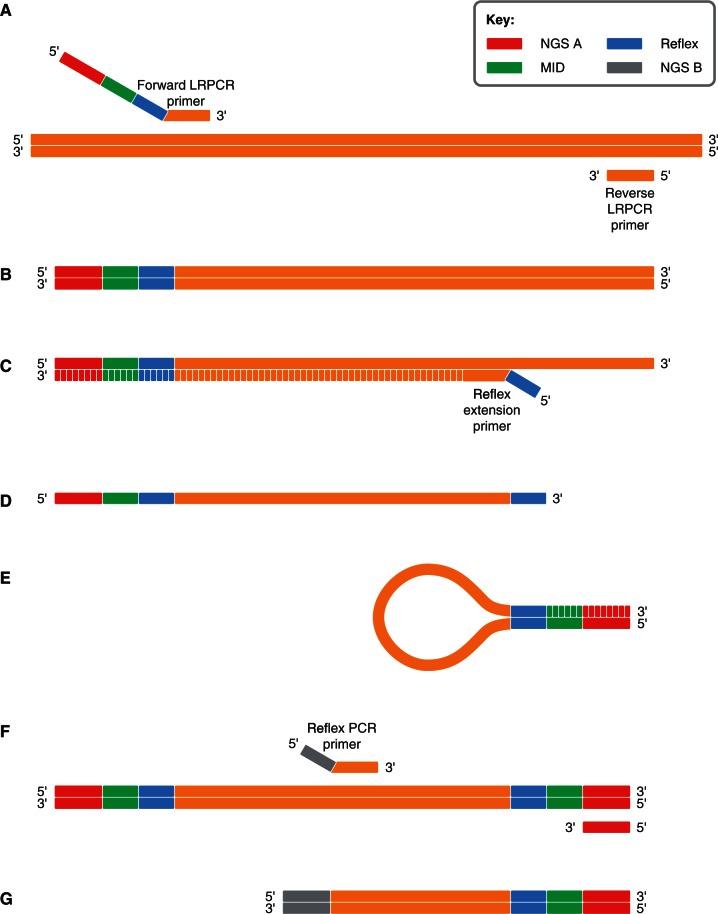
Our workflow comprises two steps, (i) LRPCR and (ii) Reflex. For LRPCR, we add a 5′-tail to the forward primer to introduce at one end of the LRPCR product, 5′–3′: a sequencing adaptor, a sample-specific MID, and a Reflex sequence. Thus, LRPCR combines enrichment and sample preparation barcoding in a single reaction (Figure 1A) and, therefore, allows us to pool, equal amounts of all such LRPCR products of each sample in the population into a single tube (Figure 1B).
Figure 1.
The Reflex workflow. The annealing positions and structure of the long-range PCR primers are shown in (A), not to scale. We generate sample-indexed long-range PCR amplicons (B), then quantify and mix them equimolarly to create a population pool. We anneal and extend a reflex-tailed extension primer (C) and extend another primer (NGS A) to reverse the polarity and generate the reflex reaction template (D). We anneal the complementary reflex sequences intramolecularly and polymerase extend to copy the MID and NGS A sequences (E) before copying back with a primer annealing at NGS A to make the material double-stranded (F). Finally, we PCR amplify the reflex products with a primer annealing at NGS A and another tailed with NGS B (shown in F) to produce sample-indexed amplicons of suitable size for NGS (G).
Instead of performing fragmentation either physically or by transposition on each individual sample, we use the Reflex step on the whole population pool to create daughter molecules tiled across the target region. The tiled series of daughter molecules are generated by adding aliquots of the population pool to individual wells of a 96-well plate for each Reflex reaction: although each Reflex reaction is a ‘single-plex’ PCR for the target, the whole population is amplified at once in each well. For each Reflex reaction, two primers are designed that flank a region of interest within the LRPCR product. The primers are spaced at an appropriate distance to satisfy the read length of a given next generation sequencing platform. First, an extension of a primer carrying a 5′ Reflex tail introduces a second inverted repeat of the Reflex sequence (Figure 1C). We then copy that strand (Figure 1D) to reverse the orientation and allow intramolecular pairing to produce a looped structure with a single-stranded 5′-tail. Polymerase extension using the 5′-tail as template then copies the cognate MID at the other end of the molecule (Figure 1E). The second primer is used in a PCR together with the sequencing adaptor primer to remove the intermediate sequence, enrich the product of the intramolecular extension and to add further sequencing adaptor domains (Figure 1F and G). All Reflex products are then quantified and again equal amounts of each pooled for sequencing. We have demonstrated we can generate such barcoded daughter Reflex products from LRPCR parent molecules as large as 10 kb.
Demonstration of Reflex workflow on CYP2D6 gene
We have used this Reflex workflow to sequence the entire CYP2D6 gene and 1 kb of upstream sequence in 95 samples. CYP2D6 is known to be a gene with variant alleles of pharmacogenomic interest (4). It also has close paralogues that makes hybridization or standard PCR approaches problematic. Selection of target-specific primers for the LRPCR, coupled with the subsequent Reflex workflow, means that we can resolve variation across the whole gene using a short-read platform. Here, we demonstrate that the Reflex workflow works with single runs on both the Roche 454 GS Junior and Life Technologies Ion Torrent PGM benchtop sequencers.
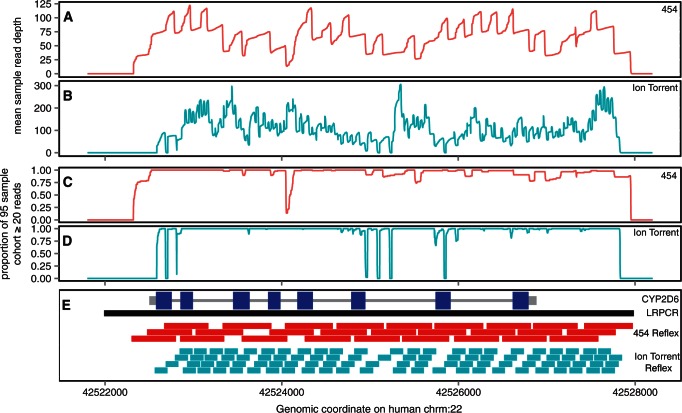
We designed two sets of overlapping daughter Reflex products across the target for both 454 and Ion Torrent platforms (Figure 2E). We amplified 95 CEU DNAs with LRPCR primers where one of the primers carries adaptor and MID sequences. We equimolarly pooled the products and divided the pool into two 96-well plates for the Reflex reactions, one for each of the desktop sequencing platforms. Reflex reactions result in individual PCR amplicons, and analysis of the concentrations of the amplicons suggests that shorter Reflex loops are formed more efficiently in general (Supplementary Figure S1). However, the relationship is not compelling and is most likely confounded by sequence context of the Reflex PCR amplicon, and/or different priming efficiencies. Concentration differences were corrected by equimolar pooling (for each NGS platform) before submitting for sequencing.
Figure 2.
Target coverage and Reflex primer design. (A and B) The mean sample read depth for each base position on the x-axis for 454 and Ion Torrent platforms, respectively. (C and D) The proportion of the 95 sample cohort sequenced to at least 20 reads for each base position on the x-axis for both NGS platforms. Amplicon designs (E) with CYP2D6 UTRs are shown thick grey bars, introns as thin grey bars and exons as thicker blue bars. LRPCR amplicon is shown as black, with 454 and Ion Torrent Reflex amplicons shown in red and turquoise, respectively.
The equimolar pooling of both the LRPCRs and Reflex products allows us to keep the representation of both samples and amplicons even for sequencing. This means the average depth to sequence all bases in all samples is kept minimal, as almost all bases are sequenced within 2-fold of the average depth (Table 1). Plotting mean base coverage across the target region indicates high uniformity (Figure 2A and B). Occasionally, Reflex primer design constraints cause a small loss in target coverage, most notably here with Ion Torrent designed amplicons (Figure 2B and E), although this is likely to improve with more experience of each NGS platform read length. We can also analyse the 95 sample population cohort coverage by applying a depth cut-off of 20 reads or more per sample and calculating the proportion of the sample population cohort that achieve this minimum depth at any base across the target gene (Figure 2C and D). These data indicate high cohort coverage across the target, which is important for population-based re-sequencing studies of this kind. In addition, most bases are called at high quality (99.8% for Roche 454 GS Junior and 94.7% for Ion Torrent). 43 of the 95 samples were examined as part of the 1000 Genomes project (5) and provide a reference from which to calculate variant calling concordance. Initial discordant results were further checked by independent Sanger sequencing validation and give final 1000 Genomes project concordances of 987/990 (99.7%) for 454 and 865/885 (97.7%) for Ion Torrent (see ‘Materials and Methods’ section and Supplementary Data).
Table 1.
Sequencing metrics
| Platform | Roche 454 GS junior | Ion torrent |
|---|---|---|
| Amplicons | 27 | 95 |
| Average depth (per bp) | 63.9 | 97.5 |
| Bases within 2-fold average depth (%) | 99.75 | 94.51 |
| Based called (%) | 99.94 | 95.80 |
| Bases called >Q30 (%) | 99.82 | 94.78 |
| Duplicate call consistency (%) | 100 | 99.99 |
| Duplicate variant consistency (%) | 100 | 98.77 |
| 1000 genomes concordance (%) | 99.70 | 97.70 |
We also examined the calls for novel variants and checked that the calls were consistent with the familial structure of the samples, i.e. that we saw a novel allele in both parent and offspring. Four checks were possible in the 454 data and three in the Ion Torrent data: all were consistent. Novel SNP calling is plagued by false-positive results; however, in the Reflex approach, we demand observations of novel SNPs in the overlapping amplicons. This approach helps remove false variation.
DISCUSSION
The Reflex workflow described here is a useful technique for sequencing large numbers of samples in depth across a contiguous target. The total number of reactions is related to the sum of the number of samples and amplicons, rather than the product, as is the case with brute force PCR. The ‘hands-on’ step for each sample is, therefore, a single cheap LRPCR, which is easily automated, and samples are pooled immediately after LRPCR avoiding per sample library preparation. The entire workflow can be completed in a few days. However, tiled target-specific primer synthesis is required to perform Reflex. It is therefore apt when the sample number is high in relation to the target size, as the work saving comes from amplifying the population pool at once, and the initial expense of the target-specific primers can be saved over multiple samples. Per sample reagent costs can be less than $10 when many thousands of samples are interrogated in the same contiguous region (Supplementary Table S2). Further cost reductions are also possible in the future, for example, by using a common adaptor set for multiple targets using splicing by overlap extension or ‘SOEin’ LRPCR. We believe many laboratories are interrogating the same genomic regions in many hundreds, if not thousands, of samples and would benefit from such sample-scale efficiencies. In addition, for sequencers that allow an extra indexing run (Illumina), we anticipate that this can be combined in sample batches with the Reflex MID to sequence 1000 s of samples in a single run. We have used the Reflex workflow to extract and sequence a gene target from ∼3000 human genomic DNA samples as part of an ongoing collaboration. In future, it may also be combined with molecular counting approaches (6), to generate long template reconstructions or ‘long-reads’ via the propagation of molecular identifiers across a contiguous region, achieved on a short-read NGS platform. This would help resolve haplotypes from DNA or isoforms from RNA molecules.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 2–7, Supplementary Figure 1, and additional Supplementary Data.
FUNDING
Venture Capital funding plus funding from The Wellcome Trust and the Company founders and presents independent research. Funding for open access charge: Population Genetics Technologies Ltd.
Conflict of interest statement. All authors are employed by Population Genetics Technologies Ltd., a privately financed company that develops and markets systems for genetic analysis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Shilo Dickens of the DNA Sequencing Service, University of Cambridge for the 454 GS Junior sequencing, Lindsay Patty of DeWalch Technologies for the Ion Torrent PGM sequencing and Peter Dimitrov for running the automated Reflex coverage analysis pipeline in R.
REFERENCES
- 1.Harismendy O, Frazer K. Method for improving sequence coverage uniformity of targeted genomic intervals amplified by LR-PCR using Illumina GA sequencing-by-synthesis technology. Biotechniques. 2009;46:229–231. doi: 10.2144/000113082. [DOI] [PubMed] [Google Scholar]
- 2.Knierim E, Lucke B, Schwarz JM, Schuelke M, Seelow D. Systematic comparison of three methods for fragmentation of long-range PCR products for next generation sequencing. PLoS One. 2011;6:e28240. doi: 10.1371/journal.pone.0028240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adey A, Morrison HG, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, Shendure J. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goetz MP, Kamal A, Ames MM. Tamoxifen pharmacogenomics: the role of CYP2D6 as a predictor of drug response. Clin. Pharmacol. Ther. 2008;83:160–166. doi: 10.1038/sj.clpt.6100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casbon JA, Osborne RJ, Brenner S, Lichtenstein CP. A method for counting PCR template molecules with application to next-generation sequencing. Nucleic Acids Res. 2011; 39:e81. doi: 10.1093/nar/gkr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.