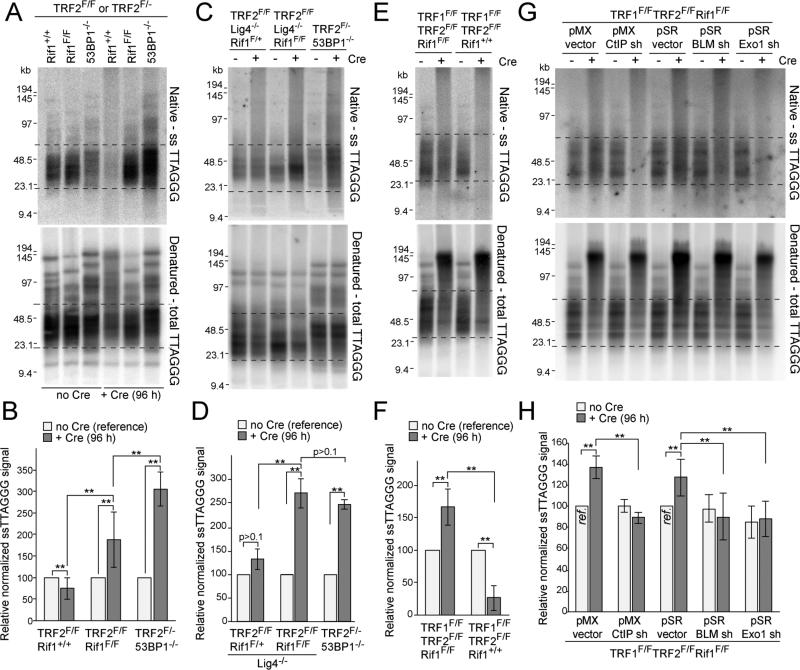
Figure 3. Rif1 blocks 5’ end resection at dysfunctional telomeres.
(A) Telomeric overhang assays on TRF2F/FRif1+/+, TRF2F/FRif1F/F and TRF2F/-53BP1-/- MEFs. Native in-gel hybridization of MboI/AluI digested DNA with end-labeled [AACCCT]4 (top) and re-hybridization with the same probe after denaturation in situ (bottom). Dashed lines represent the bulk of free (unfused) telomeres used for quantification. (B) Quantification of overhang assays as in (A). Overhang signals in no Cre samples was set at 100%. (C, D) Overhang assays on TRF2F/FRif1F/+Lig4-/-, TRF2F/FRif1F/FLig4-/- and TRF2F/-53BP1-/- MEFs and quantification as in (B). (E, F) Overhang assays on TRF1F/FTRF2F/FRif1+/+ and TRF1F/FTRF2F/FRif1F/F MEFs and quantification. (G, H) Overhang assays to measure dependency on CtIP, BLM, and Exo1 and quantification. Cells infected with either pMX or pSR with or without the indicated shRNAs and treated with Cre for 96 h. Samples with empty vectors and no Cre (ref.) were used as references. Data in (B,D,F,H) represent means of ≥3 experiments ±SDs. ** indicates p values <0.05 (two-tailed paired Student's t-test). MEFs are SV40-LT immortalized.