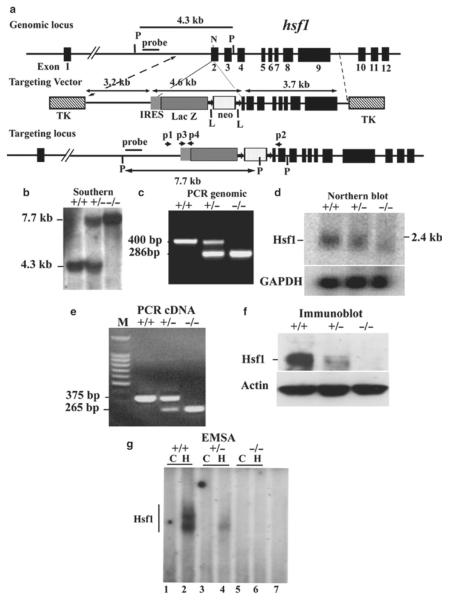
Fig. 3.
Targeted disruption of hsf1 by homologous recombination. (a) Schematic of segment of the hsf1 locus, targeting construct, and targeted locus. Coding exons are boxes in black, beginning with exon 2 (12). Targeting construct replaces the coding region for 55 bp of exon 2. LoxP flanked, PKG-neomycin cassette with upstream IRES-LacZ is indicated. Two TK genes were used for negative selection. “Outside probe” was used for screening ES cell clones to distinguish between endogenous and targeted alleles. The 3.2 and 3.7 kb fragments are the proximal and distal hsf1 homologous segments in the targeting vector (11). The final insert between the two TK genes is 13 kb. N and P represent NheI and PstI sites, respectively. Primers p2, p2, p3, and p4 are used for genotyping. (b) Southern blot analysis of genomic DNA prepared from tails of wild-type (+) and targeted mutant mice (−). The 7.7 and 4.3 kb bands are PstI-digested fragments corresponding to the targeted (−) and wild-type (+) alleles, respectively. (c) PCR analysis of tail DNA derived from wild-type (+) and targeted mutant mice (−) showing 420 and 890 bp fragments derived from wild-type and targeted alleles, respectively. (d) Northern blot analysis of total RNA derived from mouse embryo fibroblasts (MEFs) of wild-type (+) or mutant (−) mice. Full-length murine hsf1 cDNA was used as a probe. Hsf1 generates an approximately 2.4 kb fragment. GAPDH is shown for equal loading. (e) cDNA from wild-type (+) and hsf1 mutant (−) mice were amplified using forward primer located in exon 1 and reverse primers located in exon 3. Sequencing the 375 bp (+) and 265 bp (−) fragment indicated normal splicing of exons 1, 2, and 3 and splicing of exon 1 to exon 3, respectively. (f) Immunoblot analysis of extracts of MEFs derived from wild-type (+) or mutant (−) hsf1 analyzed by SDS-PAGE using antibody to Hsf1. Actin is shown as an indicator of loading. (g) Electrophoretic mobility shift assays (EMSAs) (11). Nuclear extracts of wild-type (+) or hsf1 mutant (−) MEFs were prepared from untreated control (C) or heated (43°C for 20 min plus 30 min recovery at 37°C to ensure Hsf1 activation). Lanes 1 and 2, 3 and 4, 5 and 6 represent untreated control (C) and heated (H) samples, respectively. Lane 7 is the same extract as in lane 2, but with 200× excess cold HSE to show specificity.