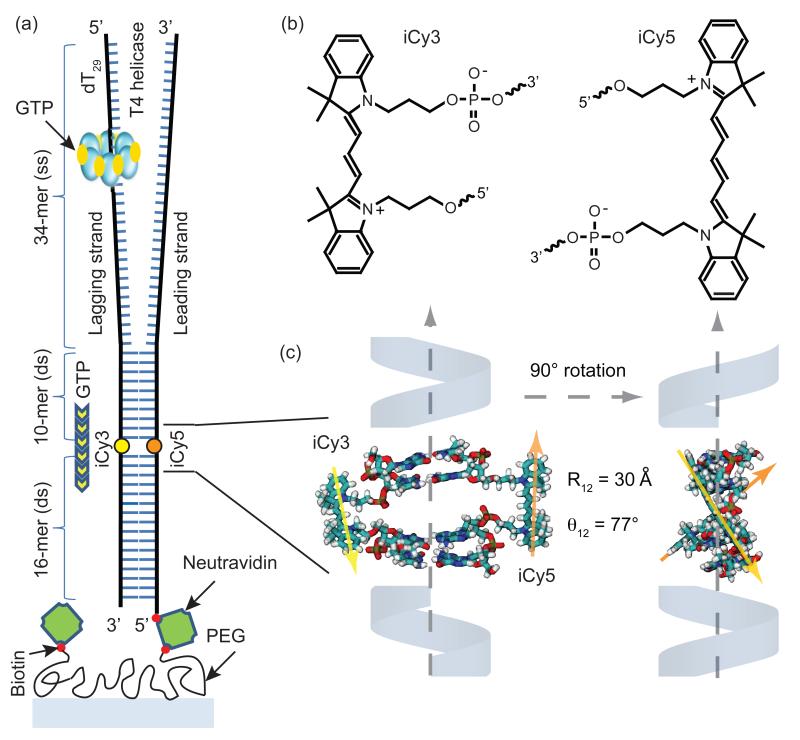
Figure 1. The DNA replication fork construct used in this work.
(a) The T4 helicase binds to the d(T)29 loading sequence on the lagging strand, and unwinds the double-stranded (ds) region in the presence of GTP. The strands within the dsDNA region are internally labeled with the FRET donor-acceptor chromophores iCy3 and iCy5, respectively. (b) The donor-acceptor iCy3/iCy5 chromophores are incorporated into the sugar-phosphate backbone using a phosphoramidite oligonucleotide synthesis procedure (see text). (c) Molecular model showing the three-dimensional structure of the iCy3/iCy5 labeled duplex region of the DNA constructs. The chromophores are rigidly positioned within the sugar-phosphate backbone with transition dipole center-to-center distance R12 ~ 30 Å and relative angle θ12 ~ 77°. When the primosome complex unwinds the duplex the distance between the donor and acceptor chromophores becomes greater than the Förster distance (~ 50 Å), disrupting the FRET efficiency and leading to a decrease in acceptor emission and a concomitant increase in donor emission.