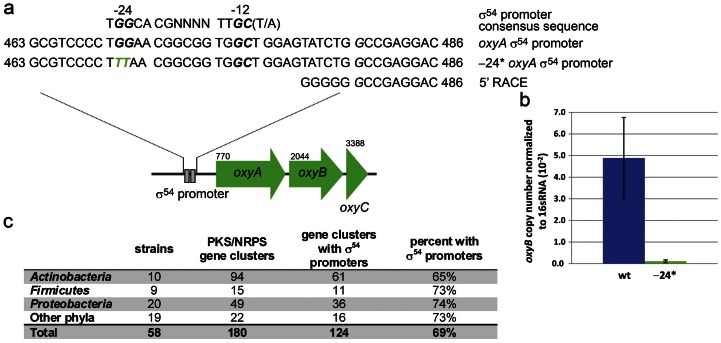
Figure 3. Putative σ54 promoters are found in the majority of polyketide and non-ribosomal peptide biosynthetic gene clusters.
(a) The oxyABCDEF operon from the S. rimosus oxytetracycline biosynthetic pathways contains a putative σ54 promoter. The promoter shows high homology to the σ54 promoter consensus sequence especially at the key −12 and −24 positions. A mutation of the highly conserved GG residues at −24 (−24*) and 5′ RACE were used to confirm that this promoter is responsible for direct, positive transcriptional regulation of the oxyB gene by σ54 over-expression. (b) Transcription of the oxyABCDEF operon is directly controlled by the σ54 promoter upstream of oxyA. Mutation of the highly conserved GG residues −24 from the transcriptional start site to TT leads to a greater than 40 fold decrease in oxyB transcript levels as determined by qPCR. qPCR data was obtained from BAP1 transformed with pDCS11 and either pDCS61 or pDCS62. (c) Results of a bioinformatics analysis of 58 bacterial genomes containing 180 polyketide synthase (PKS) and non-ribosomal peptide synthetase (NRPS) biosynthetic gene clusters show that the majority of gene clusters contain putative σ54 promoters. It is particularly intriguing that Actinobacteria possess gene clusters with σ54 promoters since they lack the gene encoding σ54. See also Tables S2, S3, S4.