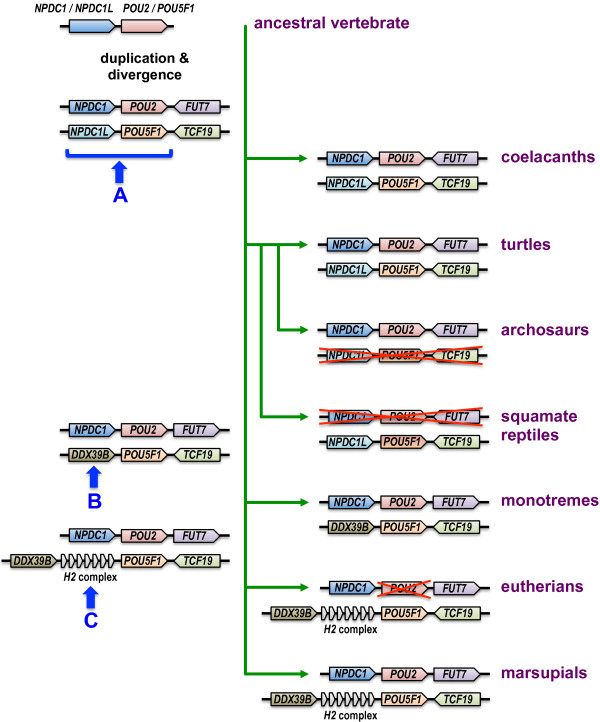
Figure 2.
Synteny at the POU5F1 and POU2 loci in sarcopterygians. The synteny of POU5F1 and POU2 with other genes in extant species is shown on the right-hand side. Proposed ancestral genomic rearrangements that explain the current synteny are shown on the left-hand side. (A) A multigenic duplication in a sarcopterygian ancestor gave rise to POU5F1 and NPDC1L, causing POU5F1 to flank TCF19 and POU2 to flank FUT7. (B) In a common ancestor of mammals, deletion of NPDC1L caused POU5F1 to flank DDX39B. The orientation of FUT7 also was inverted. (C) In a common ancestor of therian mammals, the H2 major histocompatibility complex became inserted between POU5F1 and DDX39B.